Method and kit thereof for detecting BRAF gene mutation
A kit and gene technology, applied in the field of a method for detecting BRAF gene mutation and its kit, can solve the problems of melting temperature drop, lack of specificity, sensitivity and accuracy, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
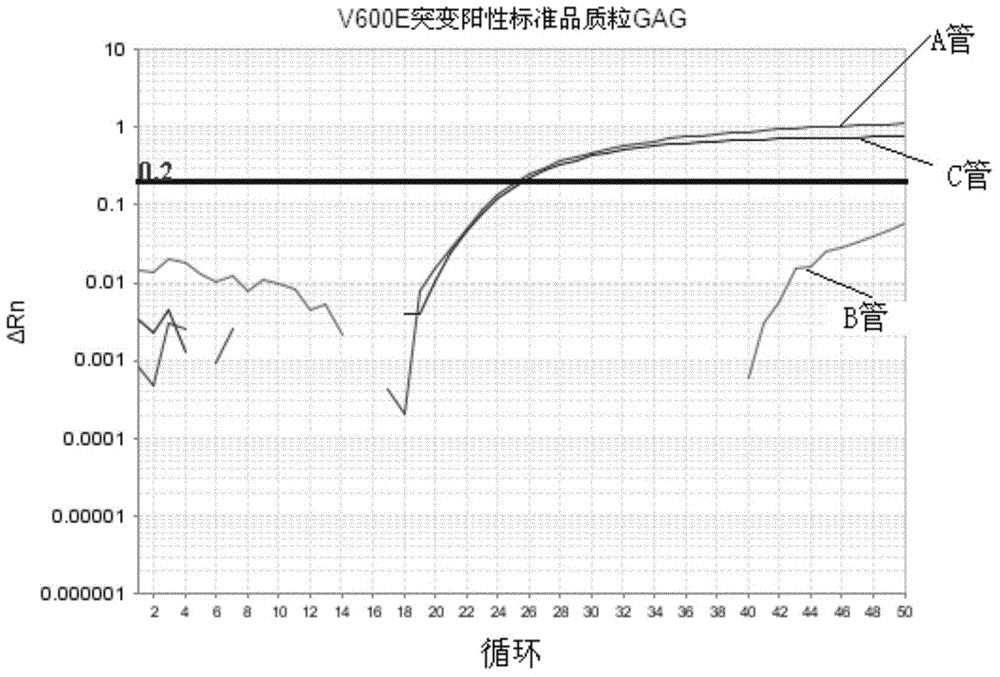
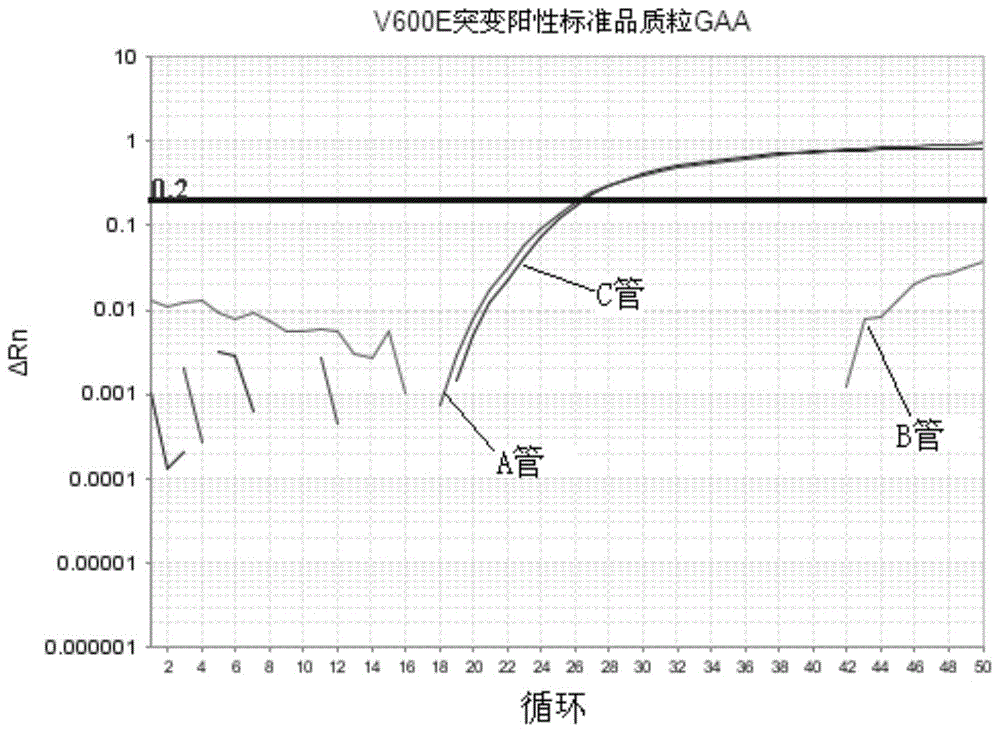
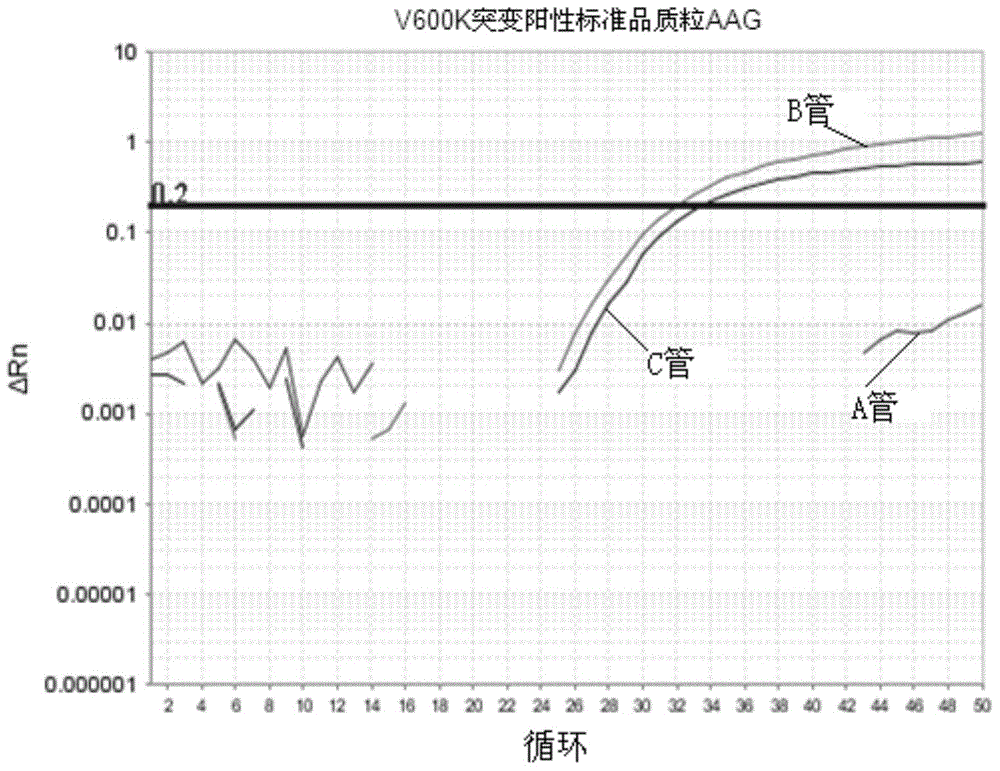
[0082] Embodiment 1 is directed at the detection of V600E and V600K mutant plasmid standards
[0083] V600E contains two nucleotide mutation sequences: GAG and GAA
[0084] Main source of reagents:
[0085] 1.1 Designing PNAs
[0086] PNA was synthesized by Panagene in South Korea and PNA Bio Inc. in the United States.
[0087] PNA-NBN-W:NH 2 -TTGGTCTAGCTACA NBN AAATC-COOH; wherein, N represents A, C, G or T, and B represents C, G or T; the described merged sequences are all mixed at a ratio of 1:1.
[0088] PNA-HNN-K: NH 2 -TTGGTCTAGCTACA HNN AAATC-COOH; where, H means A, C or T, N Indicates A, C, G or T; the described merged sequences are all mixed 1:1.
[0089] PNA-BNN-E1E2:NH 2 -TTGGTCTAGCTACA BNN AAATC-COOH; where,B means C, G or T, N Indicates A, C, G or T; the described merged sequences are all mixed 1:1.
[0090] 1.2 Design of Tagman MGB probe
[0091] The Tagman MGB probe was synthesized by Yingwei Jieji (Shanghai) Trading Co., Ltd.
[0092] Probe 1 (...
Embodiment 2
[0138] Example 2 Detection of V600E and V600K mutations in colorectal cancer tissue samples
[0139] 2.1 Specimen collection:
[0140] Collect surgically resected fresh colorectal cancer tissue samples from 8 cases of cancer (the diagnosis of cancer is based on clinical and pathological diagnosis, including pathological diagnosis), and the tissue is about 1mg.
[0141] 2.2 DNA extraction method:
[0142]Select a commercially available Qiangen Genomic DNA Extraction Kit, perform genomic DNA extraction according to the instructions, dissolve the DNA with 100 μl TE, and store in -80°C in aliquots.
[0143] 2.3 DNA concentration determination
[0144] Concentrations were measured using a NanoDrop ND-1000 full-wavelength UV / visible scanning spectrophotometer.
[0145] 2.4 Specimens for V600E and V600K mutation detection
[0146] Take 125ng of sample DNA, and perform ABC three-tube detection according to method 1.6 in Example 1.
[0147] 2.5 Result Analysis
[0148] Refer to 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com