Automatic fast segmenting method of tumor pathological image
A technology for pathological images and tumors, which is applied in the field of image processing and can solve the problems of high algorithm complexity, difficulty in accurate segmentation, and long processing time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
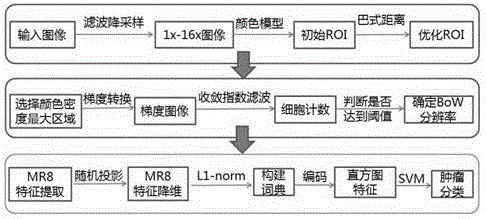
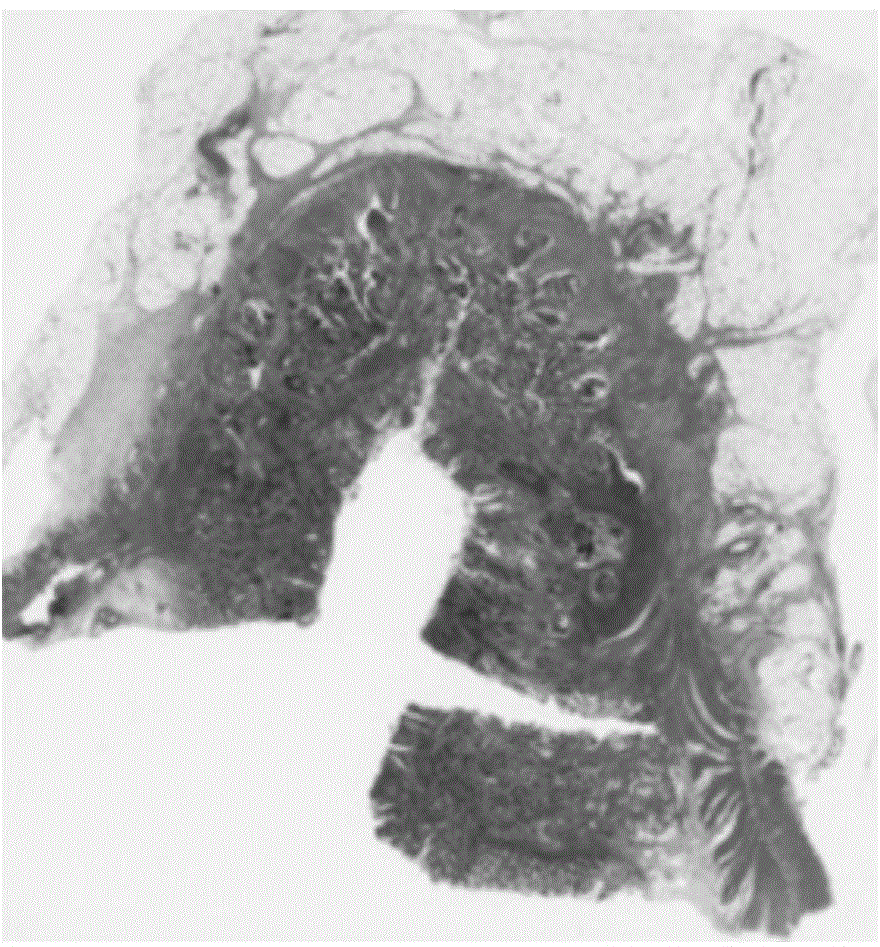
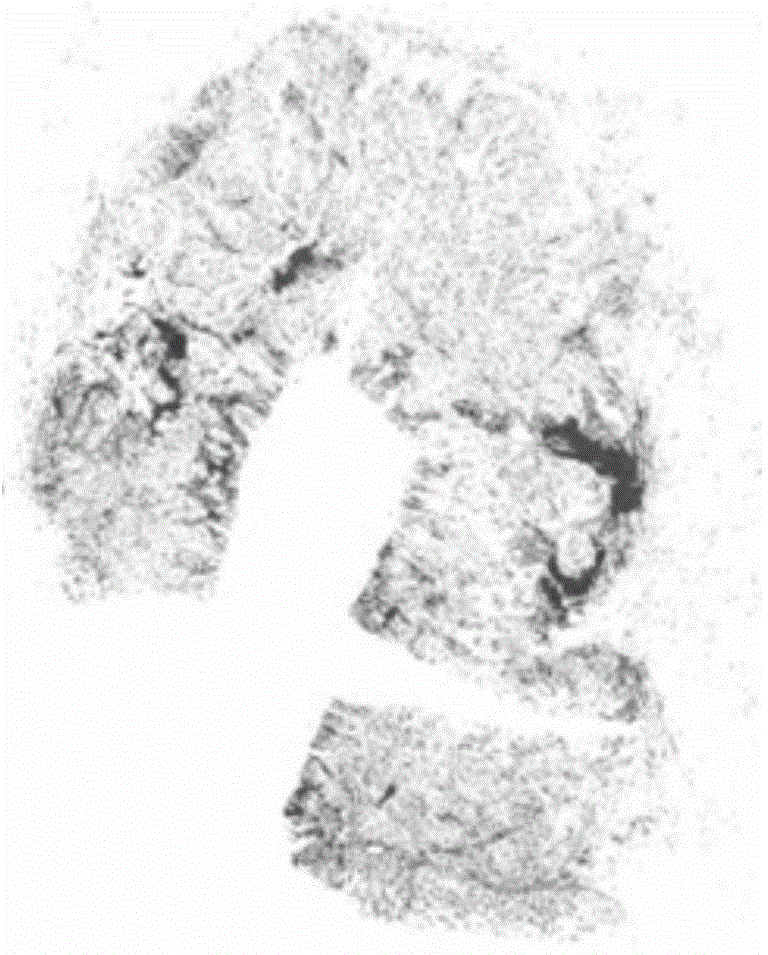
[0058] An accurate, fast and automatic segmentation method for tumor pathological images. In order to combine clinician's experience and knowledge with image processing technology, firstly, Gaussian pyramid algorithm is used to filter the original tumor pathological images, and the results are respectively obtained from 1 times, 2x, 4x, 8x, 16x pathological images, through the RGB color model and morphological "close operation" to determine the initial region of interest containing the tumor on the 1x resolution image; The iterative optimization of the initial tumor ROI was carried out from 1X resolution to 4X resolution. When the Basset distance reached a certain threshold, it was judged that the contribution of the RGB color model to the tumor ROI had been reduced to zero. Then use the convergence index filter algorithm for adaptive high-resolution selection of depth-accurate segmentation, so as to perform further segmentation at the most suitable high-resolution; finally, us...
Embodiment 2
[0091] The method in Embodiment 1 is compared with the VZ_MR8 and TFISF methods in terms of encoding time. (Both VZ_MR8 and TFISF methods are obtained by referring to the paper "Effective texture classification by texton encoding induced statistical features") KTH_TIPS is an open source database of texture images (available at www.nada.kth.se / cvap / databases / kth-tips / ) , including 10 types of images, each type of image includes 81 pictures, and each image size is 200×200 pixels. The Medical block database is based on 120 colorectal tumor pathological slice images according to the manual identification of clinicians, randomly extracting 1000 8-fold and 16-fold patches each, and each patch is 200×200 pixels in size, including 500 8-fold tumor patches, 8 times normal tissue patches500, 16 times tumor patches500 pieces, 16 times normal tissue patches500 pieces. The experimental results are the average of 20 experiments, as shown in Table 1. The encoding time based on the random p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com