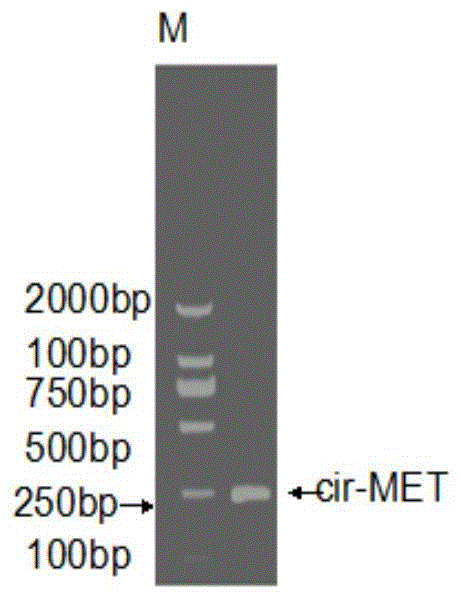
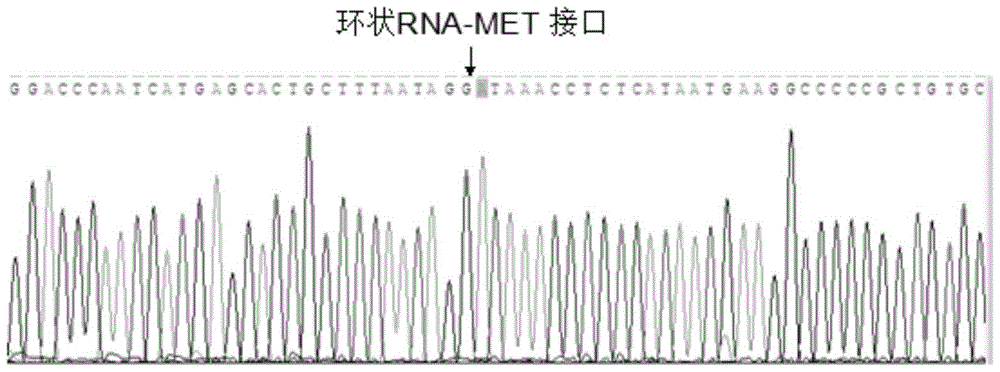
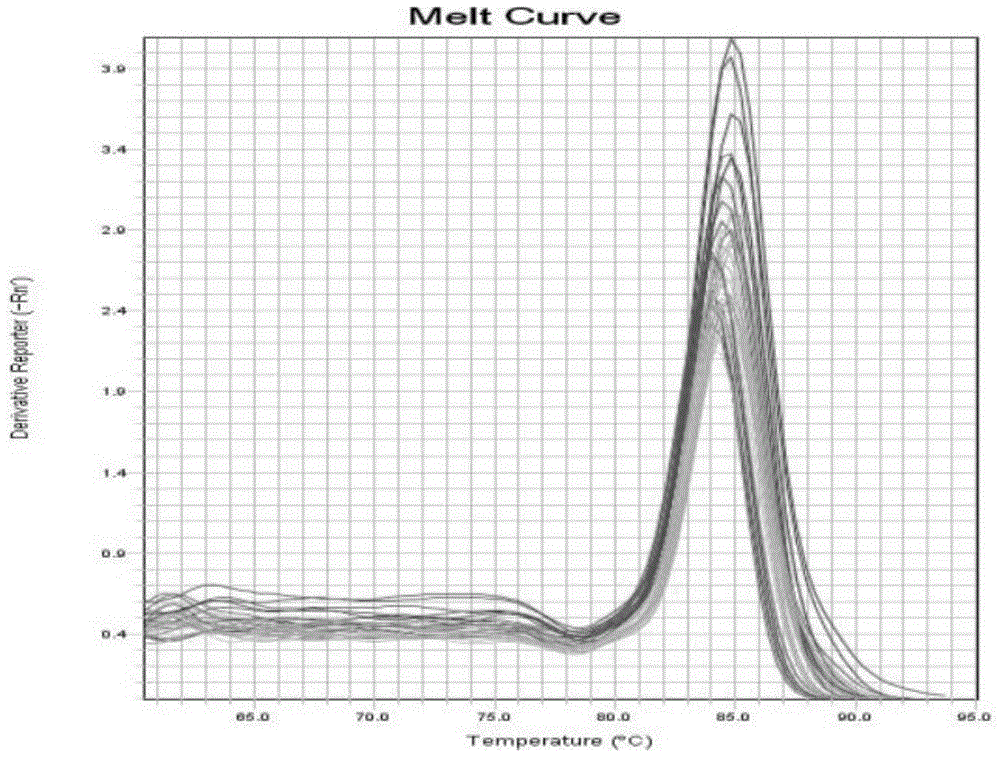
Circular RNA-met gene in liver cancer and its preparation method and fluorescent quantitative PCR detection method
A fluorescent quantitative detection, circular technology, applied in the direction of DNA preparation, biochemical equipment and methods, DNA / RNA fragments, etc., can solve the problem of no further research and achieve high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0063] A method for preparing circular RNA-MET gene in liver cancer, the preparation method comprising the following steps:
[0064] 1) Extracting total RNA in cells and tissues: extracting total RNA in liver cancer cells and liver cancer tissues;
[0065] 2) Removing the remaining genomic DNA in the extracted RNA: add the reaction solution to the total RNA extracted above, and remove the remaining genomic DNA after digestion and inactivation;
[0066] 3) Reverse transcribe RNA into cDNA: add the above RNA into a PCR tube as a template, use random reverse transcription primers, and reverse transcribe into cDNA in a PCR system;
[0067] 4) Primer design and PCR amplification, cloning and sequencing
[0068] Design PCR amplification primers, the primer sequence is MET-F: 5'CCAGATTCTGC CGAACCAATG 3', MET-R1: 5'GCTCCTCTGCACCAAGGTAA 3'; select the β-actin gene as the internal reference gene, as the correction gene of the fluorescence quantitative result data, the sequence is as fo...
Embodiment 1
[0098] A method for preparing circular RNA-MET gene in liver cancer, comprising the following steps
[0099] 1) extract the total RNA in cells and tissues, according to The Reagent reagent operating instructions extract the total RNA in liver cancer cells and liver cancer tissues; the detailed steps of extraction are:
[0100] a) Take 2 mg of liver cancer tissue, grind the tissue with liquid nitrogen until pulverized, transfer it to a 1.5ml centrifuge tube, add 1ml trizol; take about 1 million cells of liver cancer cells, add 1ml trizol;
[0101] b) Add 200 μL of chloroform, shake vigorously for 15 seconds, and let stand at room temperature for 15 minutes;
[0102] c) Centrifuge at 12000g for 15min at 4°C, the solution is divided into three layers, the RNA is dissolved in the water phase, and the water phase is transferred to another new RNase free EP tube;
[0103] d) Add 1 volume of isopropanol, vortex and mix thoroughly;
[0104] e) Centrifuge at 12000g for 10min at 4°C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com