Application of aptamer in recognition of L selectin and combination with L selectin
A nucleic acid aptamer and selectin technology, applied in the fields of biotechnology and clinical medicine, to achieve the effect of strong specificity and high affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
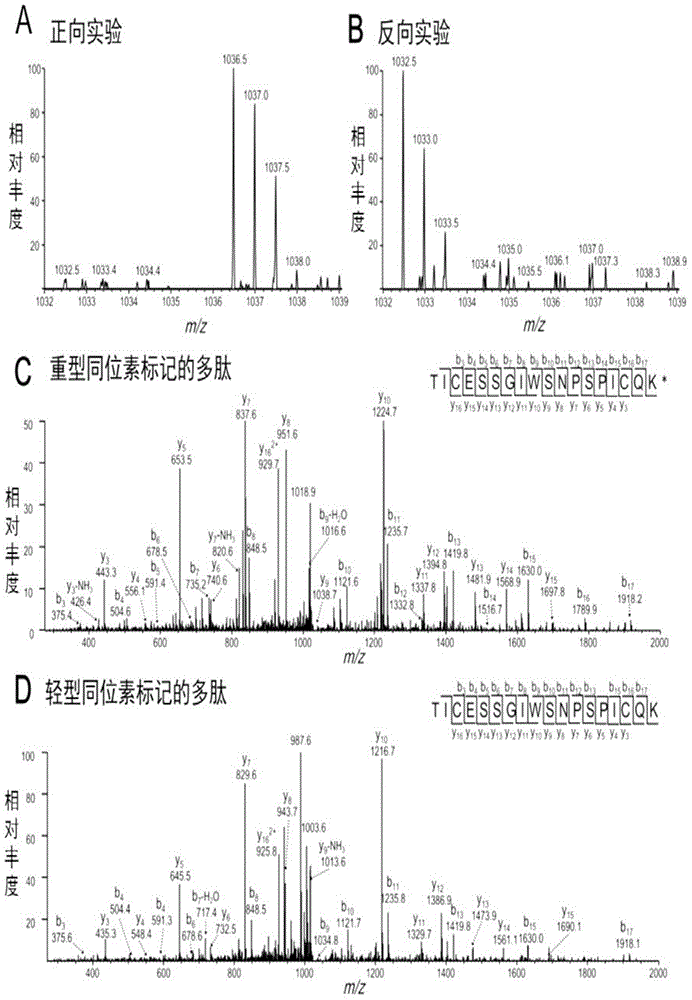
[0059] Example 1, Identification of nucleic acid aptamer Sgc-3b specifically recognizing and binding to L-selectin
[0060] 1. Preparation of nucleic acid aptamer Sgc-3b and its derivatives
[0061] 1. Synthesis of nucleic acid aptamer Sgc-3b
[0062] The nucleic acid aptamer Sgc-3b is synthesized by a DNA synthesizer, and the nucleotide sequence of the nucleic acid aptamer Sgc-3b is as follows: 5'-CTTATTCAATTCCCGTGGGAAGGCTATAGAGGGGGCCAGTCTATGAATAAG-3' (sequence 1), according to the needs of the test can be in the nucleic acid aptamer Sgc- Different molecules are marked on 3b to obtain derivatives of the nucleic acid aptamer Sgc-3b. Among them, the biotin-labeled nucleic acid aptamer Sgc-3b was selected in the following example 1; the fluorescein (FAM)-labeled nucleic acid aptamer Sgc-3b was selected in other examples.
[0063] 2. DNA deprotection: after deprotection with cold ammonia, then dissolve the DNA in TEAA solution;
[0064] 3. DNA purification: Purify by PAGE or h...
Embodiment 2
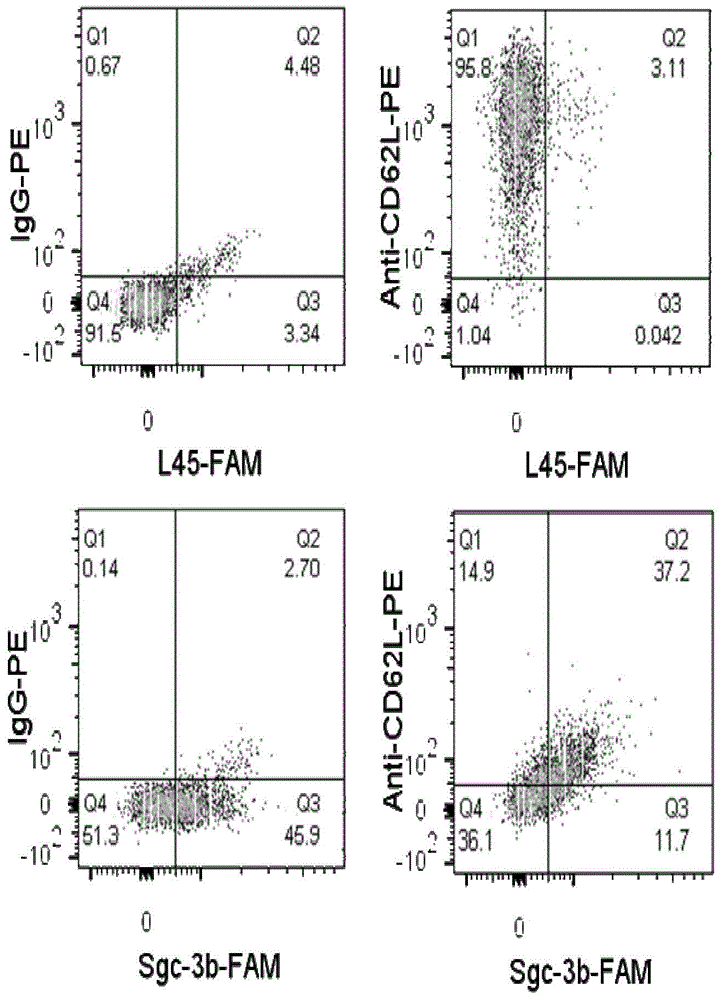
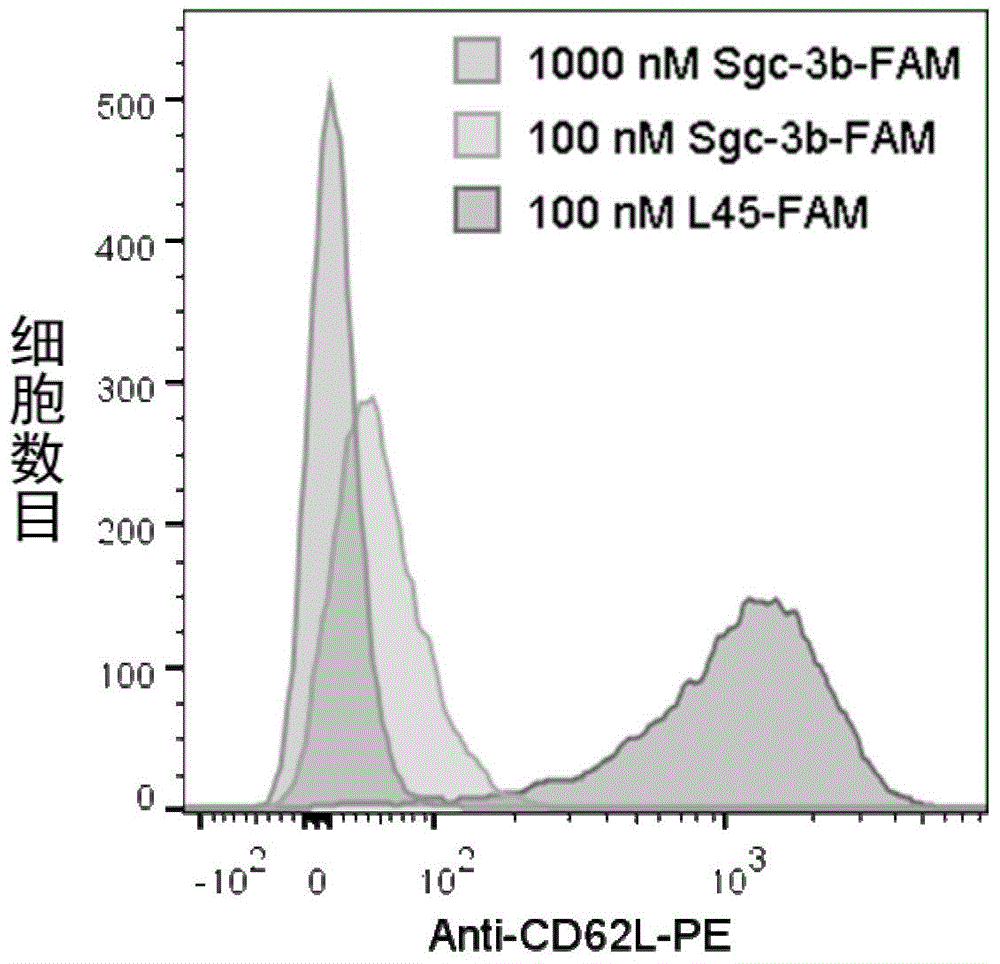
[0097] Example 2, flow cytometric analysis method to detect the binding ability of nucleic acid aptamer Sgc-3b and L-selectin
[0098] 1. Preparation of nucleic acid aptamer solution and treatment of cell lines
[0099] 1. Preparation of fluorescein-labeled nucleic acid aptamer Sgc-3b solution (Sgc-3b-FAM) (100nM)
[0100] The fluorescein-labeled nucleic acid aptamer Sgc-3b was obtained by coupling the fluorescein group to the 5' end of the nucleic acid aptamer Sgc-3b. Sgc-3b-FAM was dissolved in binding buffer, and the concentration was calibrated according to the ultraviolet absorption ( 100nM), heated at 95°C for 5min, placed on ice for 5min, and placed at room temperature for 15min.
[0101] 2. Preparation of PE-labeled anti-L-selectin antibody solution (anti-CD62L-PE)
[0102] The PE-labeled anti-L-selectin antibody is a product of eBioscience, and 0.125 μg was added to each test, with a concentration of 1.25 μg / mL.
[0103] 3. Preparation of fluorescein-labeled contro...
Embodiment 3
[0152] Example 3, Application of the nucleic acid aptamer Sgc-3b in determining the expression of L-selectin in different types of cells
[0153] 1. Preparation of fluorescein-labeled aptamer Sgc-3b solution (Sgc-3b-FAM)
[0154] Dissolve Sgc-3b-FAM in binding buffer, calibrate the concentration (100 nM) according to UV absorption, heat at 95° C. for 5 min, place on ice for 5 min, and place at room temperature for 15 min.
[0155] 2. Pretreatment of cell lines
[0156] Take one plate of each of the following 16 logarithmic growth cell lines: growing rat alveolar epithelial cells (RAEC), human embryonic lung fibroblasts (MRC-5), human alveolar epithelial cells (A549), human cervical cancer cells ( Hela), human liver cancer cells (Huh-7), human bladder cancer cells (T24), human hepatocellular carcinoma cells (SK-Hep-1), human breast cancer cells (MCF-7), drug-resistant human breast cancer cells ( MCF-7R), human ovarian cancer cells (SKOV-3), human breast cancer cells (MDA-MB-2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com