Application of an endogenous non-coding small RNA miR-873
A rnamir-873, non-coding technology, applied to the application field of endogenous non-coding small RNA, medicine, can solve the problem of little research on the regulation of target gene mRNA translation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
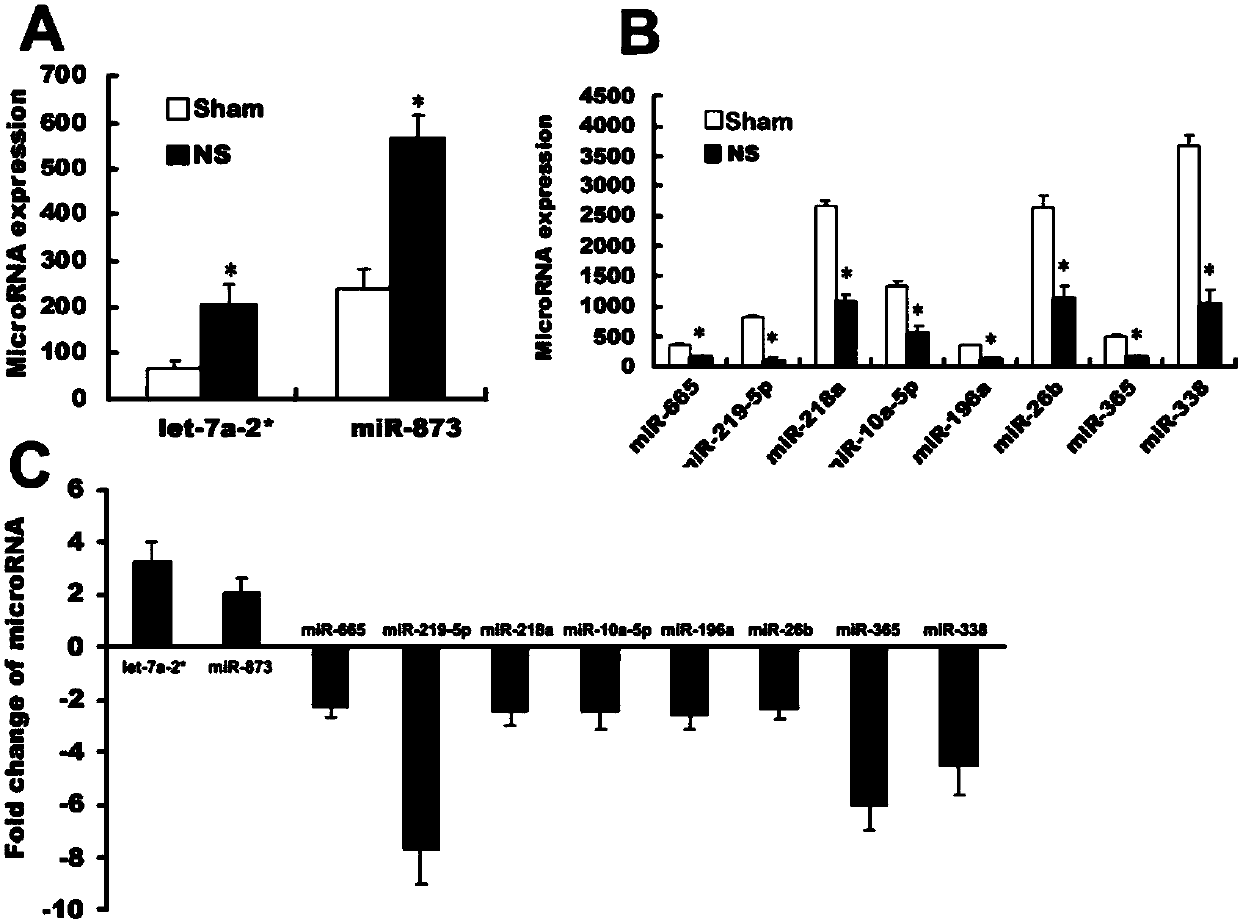
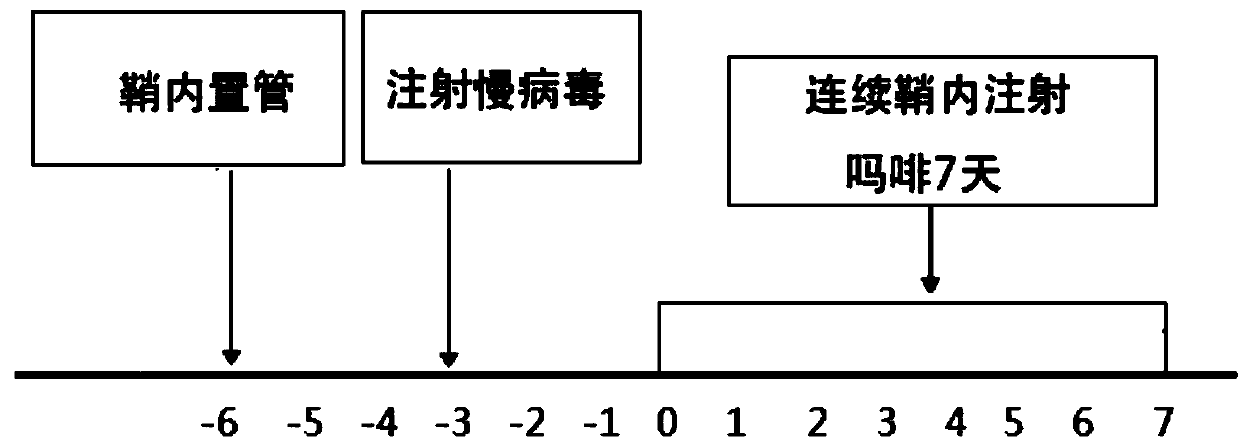
[0026] (1) To study the dynamic changes of miR-873 in the spinal cord dorsal horn and dorsal root ganglion (DRG) during the formation of morphine tolerance
[0027] Exiqon's LNA (locked nucleic acid) technology microRNA expression profile chip was used to detect chronic morphine tolerance group (NS group) and control group ( Sham group) miRNAs differentially expressed in the lumbar spinal cord dorsal horn and DRG. The pre-experiment has been completed and the microarray detection of differential expression profiles in the dorsal horn of the lumbar spinal cord on the 1st day after the formation of morphine tolerance has been completed. Using stem-loop reverse transcription primer real-time fluorescent quantitative PCR (stem-loop RT-qPCR, ABI company, TaqMan probe method) to verify the expression of miRNAs, first use miR sequence-specific stem-loop reverse transcription primer for reverse transcription, Then qPCR was performed in combination with miR-specific sense and antisens...
Embodiment 2
[0039] The specific process of lentivirus-mediated miR-873 interference vector construction:
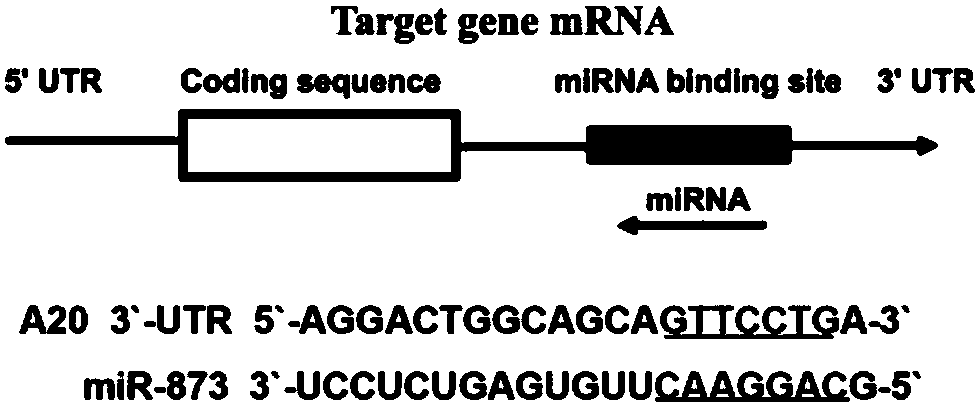
[0040] According to the miR-873 sequence, using the shRNA design program, according to the principle of RNA interference sequence design, design the interference target sequence 5'-AUAAGGAUUUUUAGGGCAUUA-3'; use EcoRI and BamHI as enzymes at both ends of the pLenR-GPH vector (Jikai Gene) Carry out enzyme digestion at the cleavage site to make it linear, perform a ligation reaction with the double-stranded interference target sequence by T4DNA ligase to form an shRNA expression vector, and transfer the ligated product into the prepared competent Escherichia coli. The clones were identified by enzyme digestion, the positive clones were selected for sequencing, and after sequencing and comparison, the positive clones were identified as the successfully constructed shmiR-873 interference vectors;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com