Determination of yeast RNA (Ribonucleic Acid) by photoluminescence of ruthenium-based metal complex
A ruthenium complex, yeast technology, applied in the field of photochemical sensing, to achieve the effect of simple quantitative detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
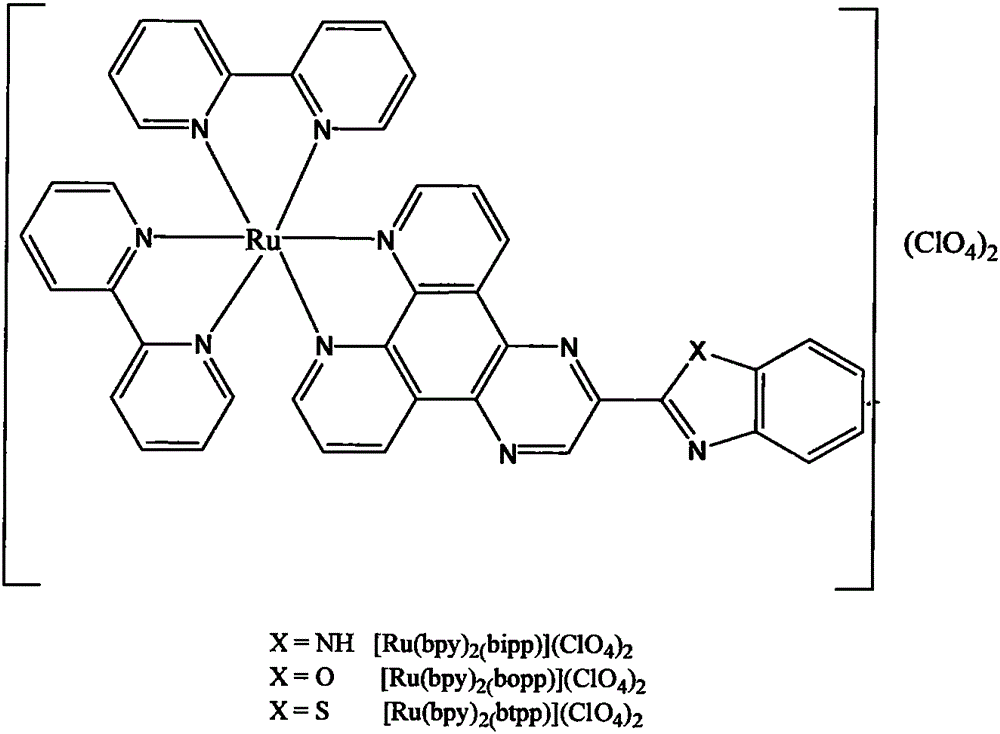
[0011] Embodiment 1: Complex [Ru(bpy) 2 (bipp)] (ClO 4 ) 2 、[Ru(bpy) 2 (bopp)] (ClO 4 ) 2 and [Ru(bpy) 2 (btpp)] (ClO 4 ) 2 Synthesis
[0012] Complex [Ru(bpy) 2 (bipp)] (ClO 4 ) 2 、[Ru(bpy) 2 (bopp)] (ClO 4 ) 2 and [Ru(bpy) 2 (btpp)] (ClO 4 ) 2 According to the method reported by our research group [Han, M.J.; Duan, Z.M.; Hao, Q.; Wang, K.Z.J.Phys.Chem.C2007, 111, 16577-16585.] Synthesis.
[0013] The ligands bipp, bopp and btpp are pressed by pyrazine[2,3-f][1,10]phenanthroline-2-carboxylic acid and o-phenylenediamine, 2-aminophenol and 2-aminothiophenol, respectively. The molar ratio of 1:1 is obtained by condensation reaction in polyphosphoric acid heated to 200°C. After the reaction was completed and cooled to room temperature, the mixture was poured into 400 mL of cold water with stirring, and the pH of the system was adjusted to 7 with concentrated ammonia water. The obtained precipitate was suction filtered to obtain a crude product, which was recrys...
Embodiment 2
[0017] Embodiment 2: Complex [Ru(bpy) 2 (bipp)] (ClO 4 ) 2 、[Ru(bpy) 2 (bopp)] (ClO 4 ) 2 and [Ru(bpy) 2 (btpp)] (ClO 4 ) 2 Determination of Yeast RNA by Photoluminescence and Drawing of Working Curve
[0018] Fluorescence emission spectra were measured on a Shimadzu RF-5301PC fluorescence spectrophotometer with an excitation wavelength of 450 nm.
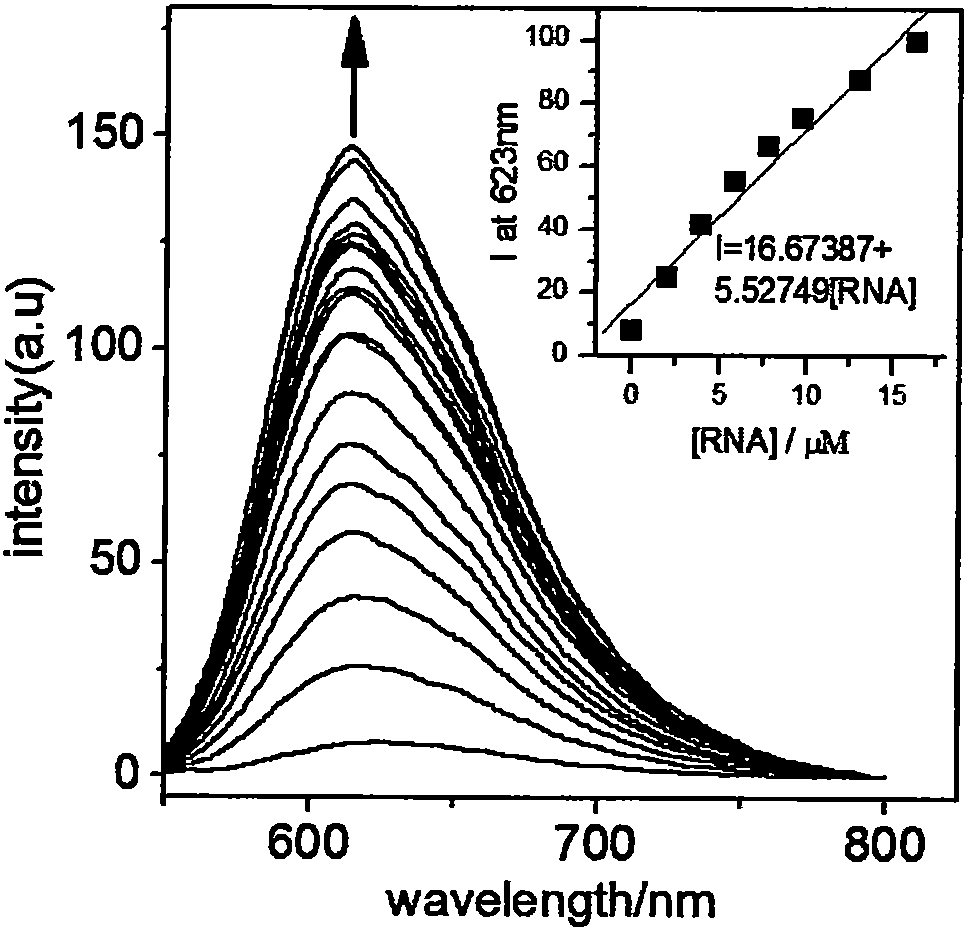
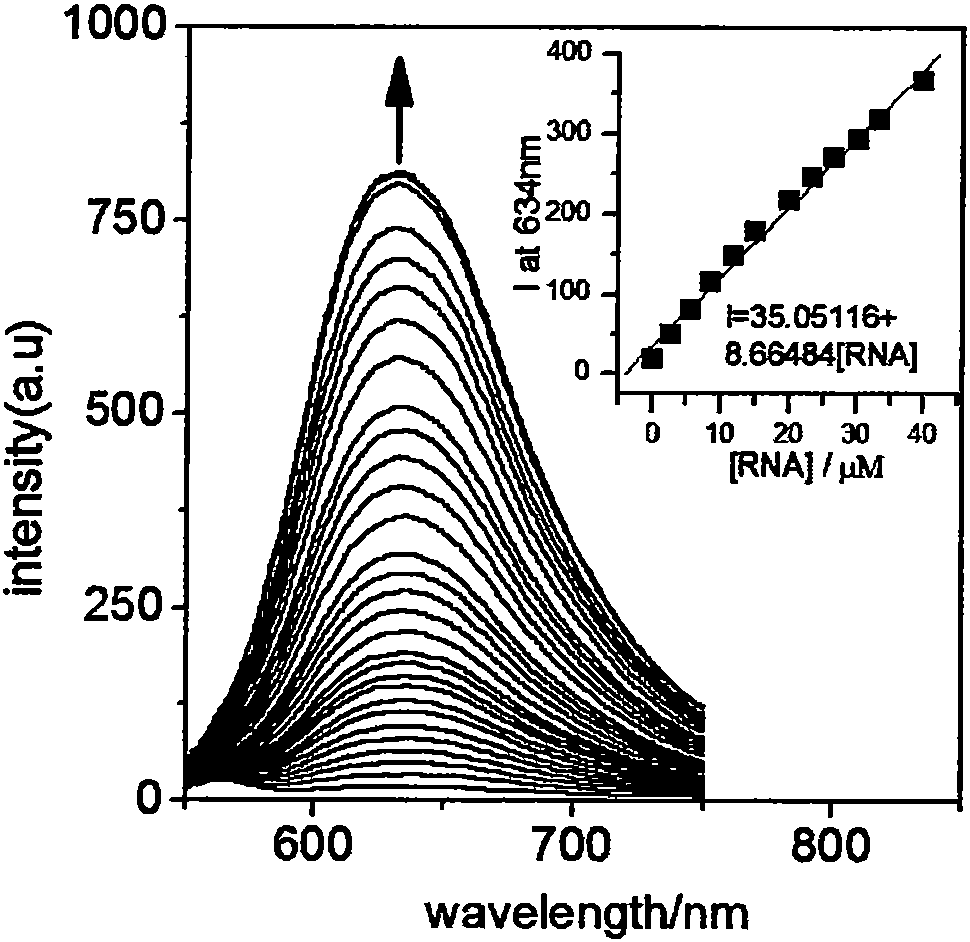
[0019] [Ru(bpy) at a concentration of 4.6 μM 2 (bipp)] (ClO 4 ) 2 The yeast RNA solution was continuously added to the Tris-HCl (5mM, pH=7.1) solution, and the change of the emission spectrum was measured with the increase of the yeast RNA concentration until the emission spectrum of the complex did not change any more. When the concentration of yeast RNA reached 160μM, the emission spectrum of the complex did not change, and the intensity of the emission peak at 623nm gradually increased with the increase of RNA concentration, accompanied by a blue shift of 9nm. Wherein the concentration of yeast RNA is in the range of...
Embodiment 3
[0022] Embodiment 3: Complex [Ru(bpy) 2 (bipp)] (ClO 4 ) 2 、[Ru(bpy) 2 (bopp)] (ClO 4 ) 2 and [Ru(bpy) 2 (btpp)] (ClO 4 ) 2 Determining the detection limit of yeast RNA
[0023] Fluorescence emission spectra were measured on a Shimadzu RF-5301PC fluorescence spectrophotometer with an excitation wavelength of 450 nm.
[0024] Take 6 copies [Ru(bpy) 2 (bipp)] (ClO 4 ) 2 (4.6 μM) Tris-HCl (5 mM, pH=7.1) solution, the emission spectra of the solutions were measured respectively, and six parallel measurement results were obtained. The luminous intensity of the complex solution at 623nm was obtained from the emission spectrum measured each time, which were 8.138, 8.039, 8.291, 8.275, 8.141, and 8.342, and the standard deviation σ of the 6 luminous intensity values was calculated 1 = 0.1064. [Ru(bpy) 2 (bipp)] (ClO 4 ) 2 Determination of the slope K of the working curve of yeast RNA 1 =5.52749μM -1 , using 3σ / K yielded a detection limit of 0.06 μM.
[0025] Take ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com