CRISPR-Cas9-specific method for knocking out porcine CMah gene and sgRNA for specifically targeting CMah gene
A specific and genetic technology, applied in the field of gene knockout and genetic engineering, can solve the problems of low recombination efficiency, tedious work, time-consuming, low efficiency, etc., and achieve the effect of high cost and long solution cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1, Selection and design of Sus scrofa (pig) CMAH gene sgRNA target sequence
[0068] The target sequence determines the targeting specificity of the sgRNA and the efficiency of inducing Cas9 to cleave the target gene. Therefore, efficient and specific target sequence selection and design are the prerequisites for constructing sgRNA expression vectors.
[0069] 1. Selection of sgRNA target sequence for CMAH gene
[0070] For the CMAH gene, the following principles should be followed in the selection of target sequences:
[0071] (1) Find the target sequence in the exon coding region of the CMAH gene that conforms to the 5'-N(20)NGG-3' rule, where N(20) represents 20 consecutive bases, and each N represents A or T Or C or G, the target sequence that meets the above rules is located on the sense strand or the antisense strand;
[0072] (2) Select the 5 exon coding region sequences near the N-terminus, the target sequence can be located in the 5 exon coding regio...
Embodiment 2
[0088] Embodiment 2, constructing the sgRNA expression vector of CMAH gene
[0089] 1. Synthesis of DNA Inserts
[0090] (1) Synthesize the forward and reverse oligonucleotide sequences designed above
[0091] The oligonucleotide sequence can be specifically synthesized by a commercial company (such as Invitrogen) according to the provided sequence. In this example and the following examples, the effect of the target sequences listed in the No. 2 and No. 4 sequences listed in Table 1 on the knockout effect of the CMAH gene was studied.
[0092] The forward oligonucleotide sequence and reverse oligonucleotide sequence corresponding to No. 2 target sequence are as follows:
[0093] CACCGTTGAGATTGGCAGCTTCGGC (SEQ ID NO: 63);
[0094] AAACGCCGAAGCTGCCAATCTCAAC (SEQ ID NO: 64).
[0095] The forward oligonucleotide sequence and reverse oligonucleotide sequence corresponding to No. 4 target sequence are as follows:
[0096] CACCGTGCCGAAGCTGCCAATCTCA (SEQ ID NO: 65);
[0097] AA...
Embodiment 3
[0133] Example 3. Obtaining a pseudotyped lentivirus expressing CMAH sgRNA
[0134] 1. Material preparation
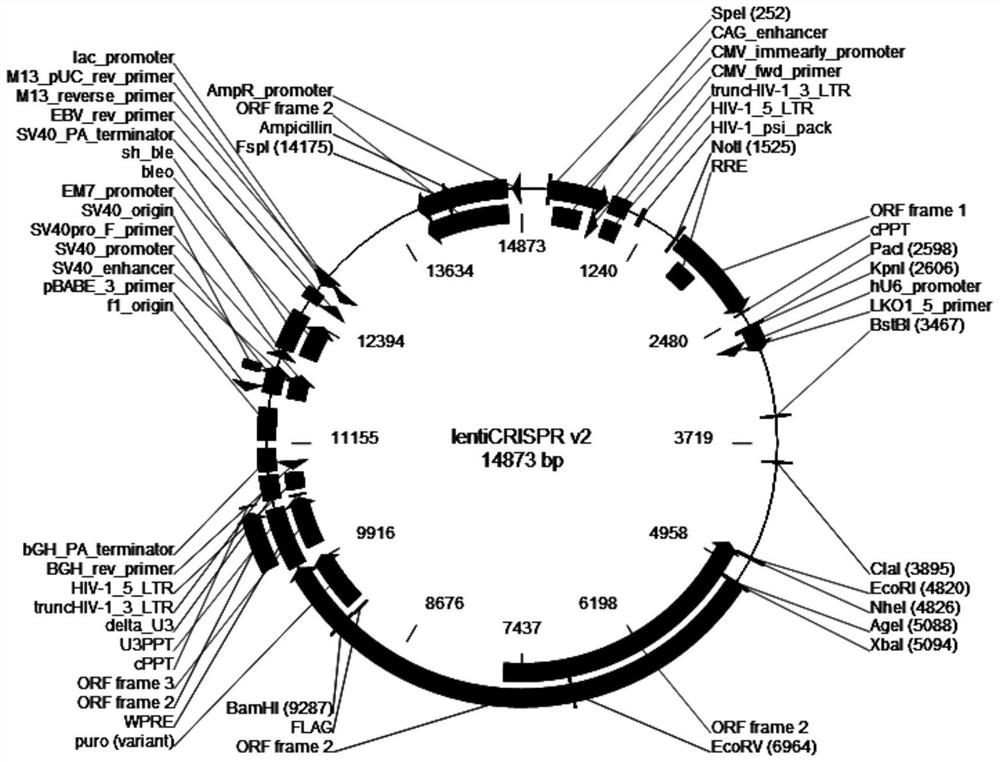
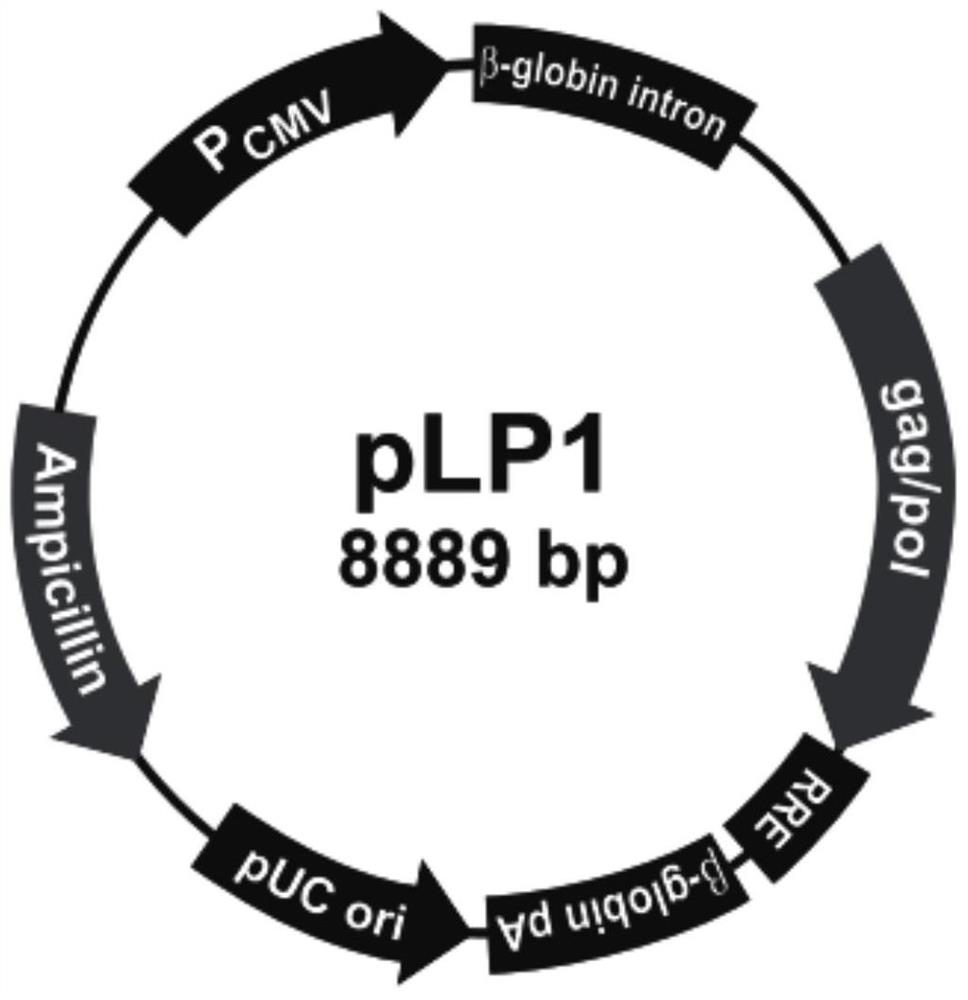
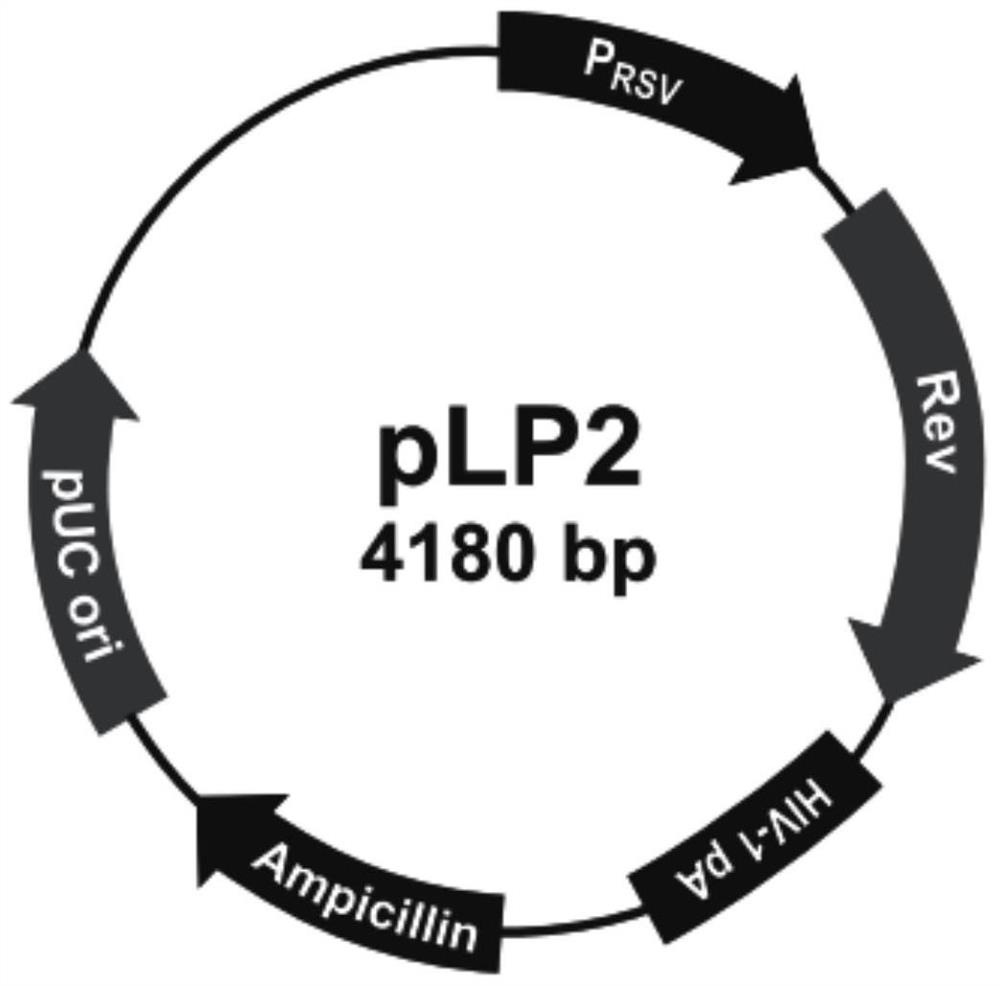
[0135] Amplify and extract packaging plasmids pLP1, pLP2, and pLP / VSVG (purchased from Invitrogen, whose maps are shown in figure 2 , image 3 and Figure 4 shown); amplify and extract vector plasmid lentiCRISPR v2-CMAH; culture packaging cell line HEK293T cells (purchased from ATCC); DMEM medium, Opti-MEM medium and fetal bovine serum FBS (purchased from Gibco); Lipofectamine2000 ( purchased from Invitrogen); HEK293T cells were cultured in 5% CO 2 In the culture environment of 37°C, the culture medium is DMEM medium containing 10% FBS.
[0136] 2. Transfection and Viral Packaging
[0137] Day 1: Passage the packaging cell line HEK293T to a 10cm dish, about 30% confluence;
[0138] The next day: when HEK293T reaches 80% confluence, transfect according to the following recipe:
[0139] Prepare Mixture 1, containing:
[0140] lentiCRISPR v2-CMAH: 6 μg
[0141] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com