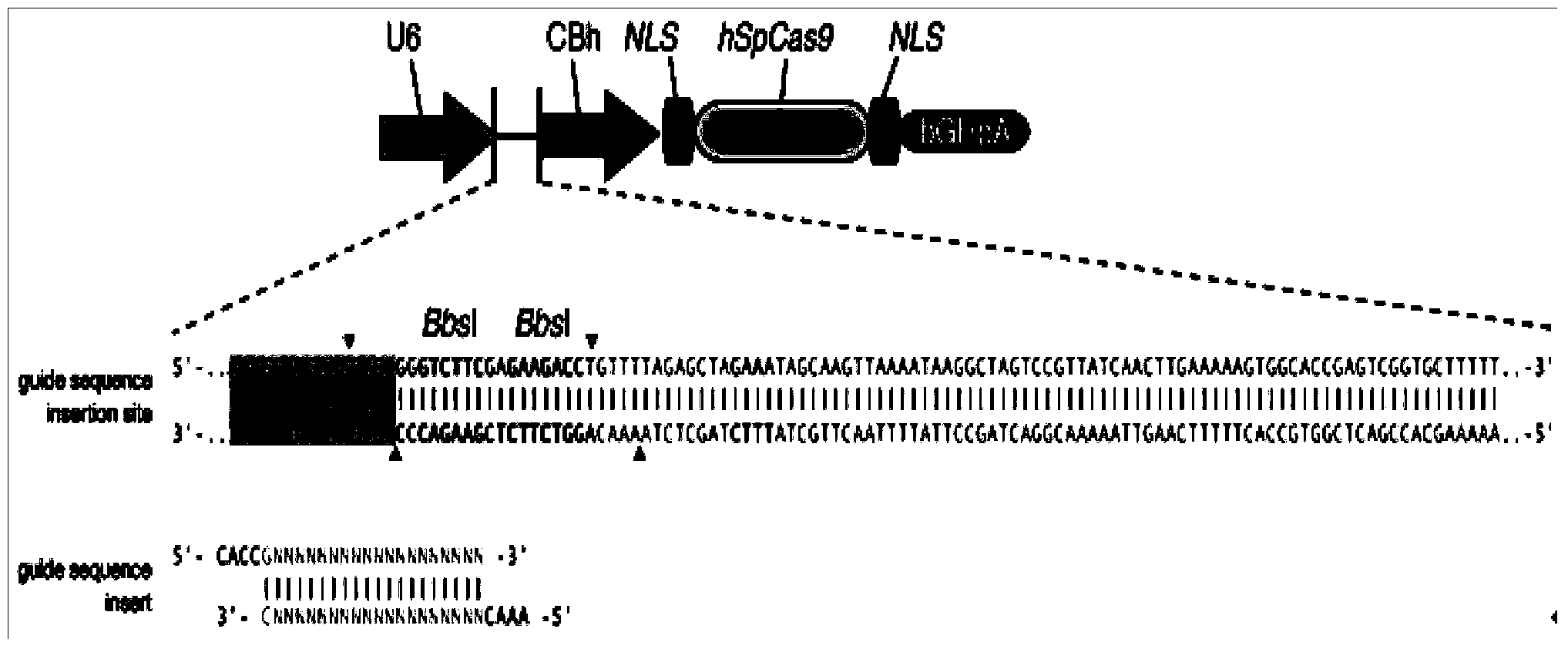
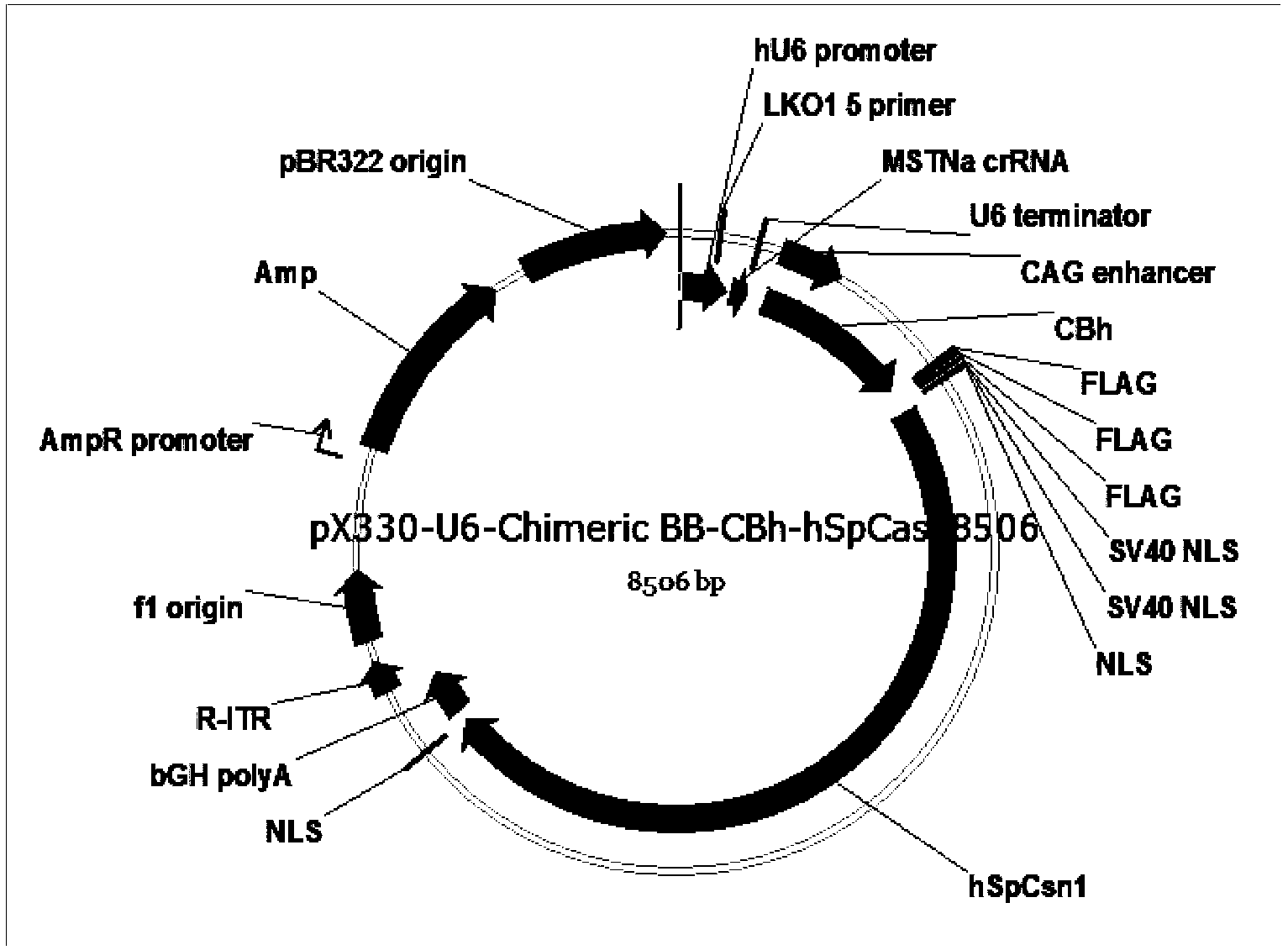
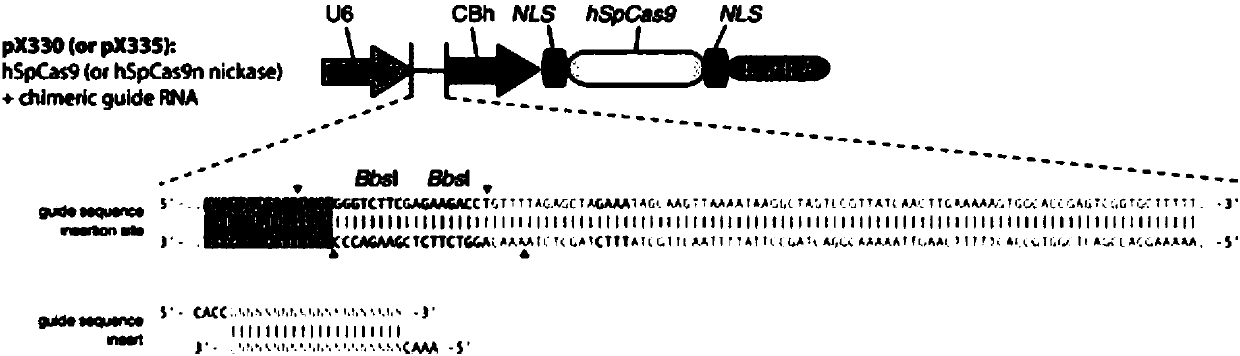
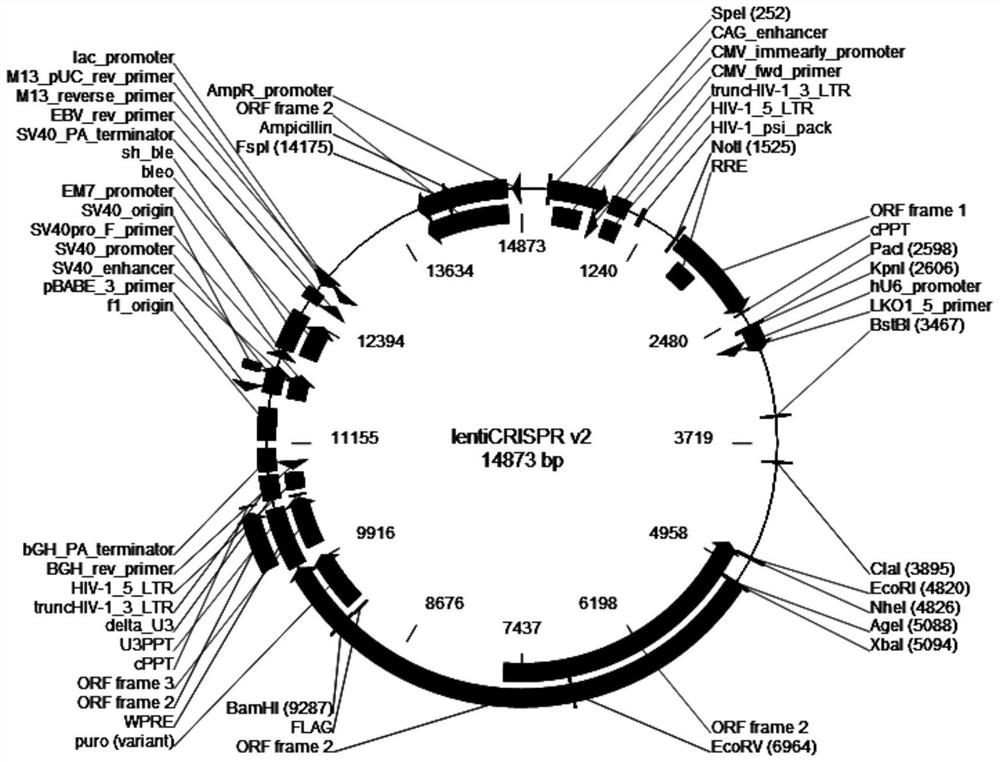
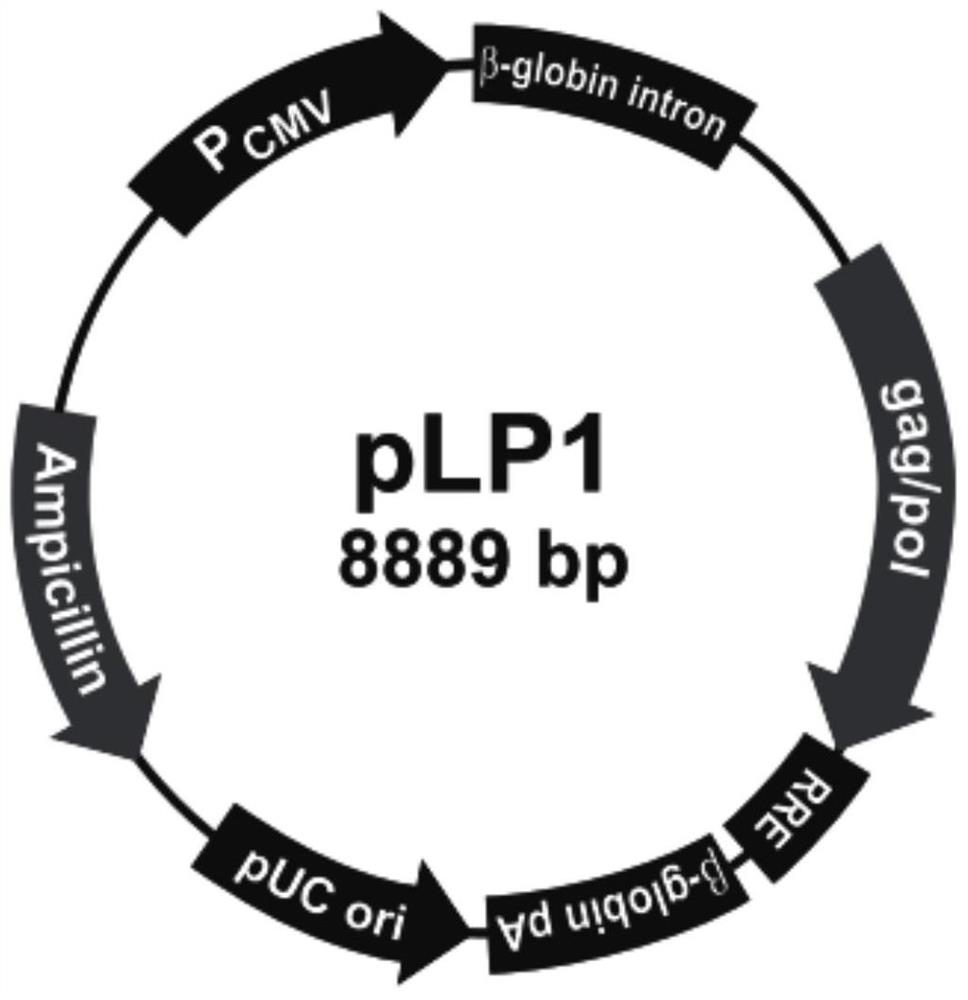
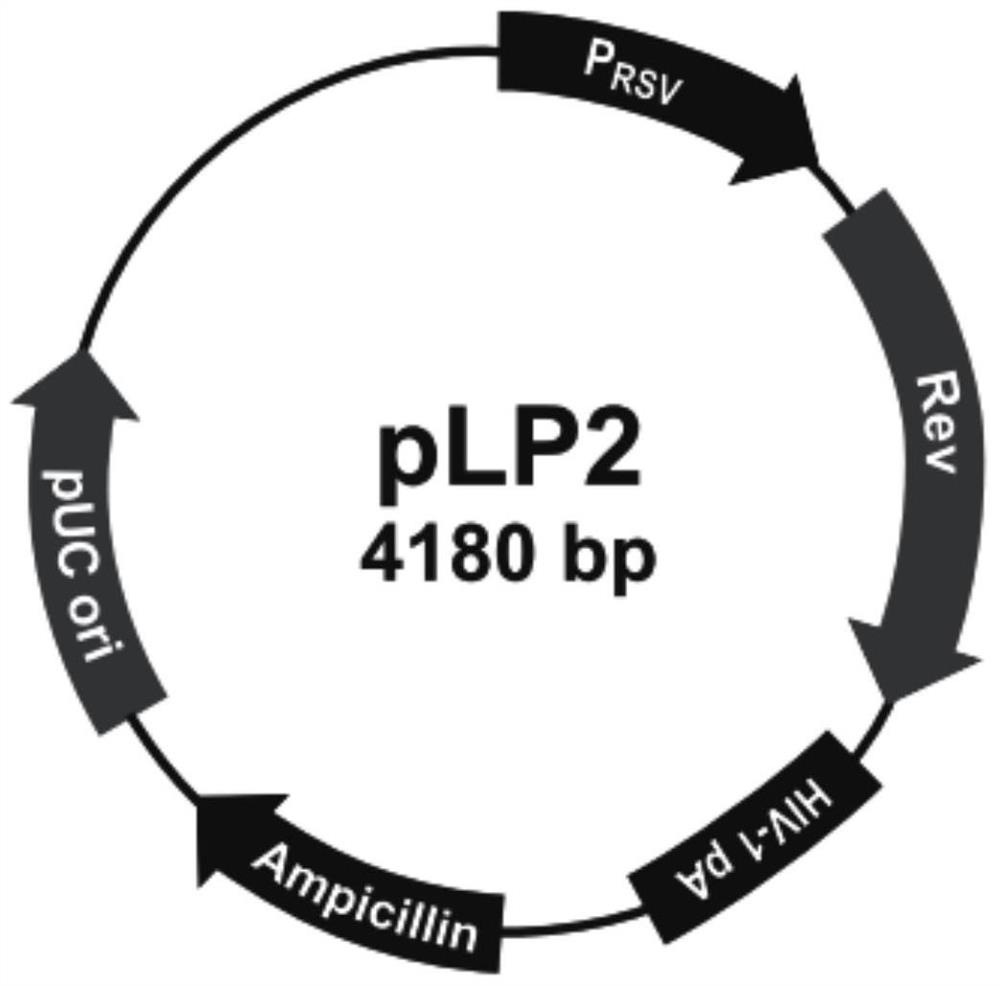
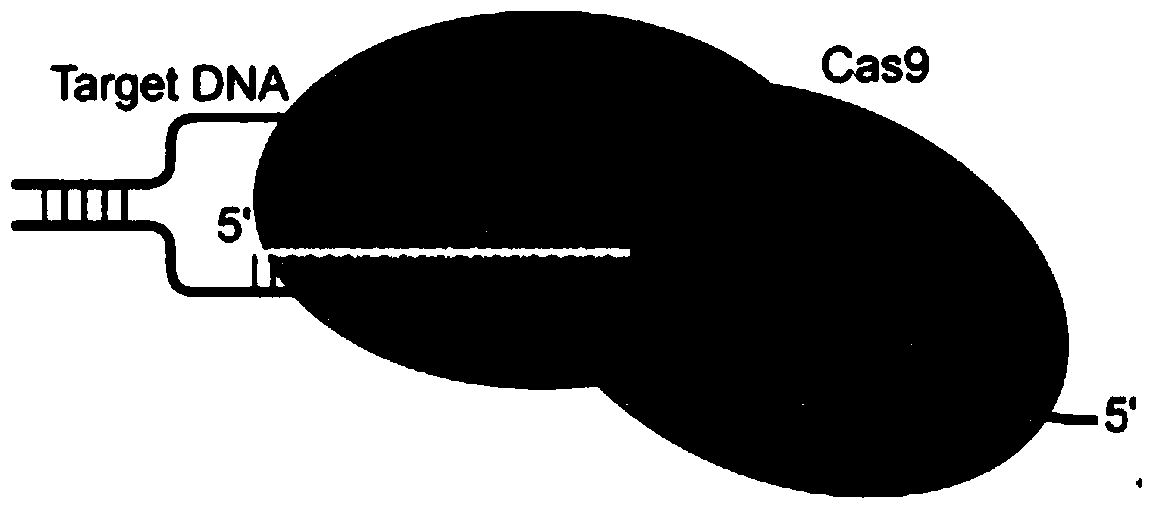
Patents
Literature
33results about How to "Knockout Efficient" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
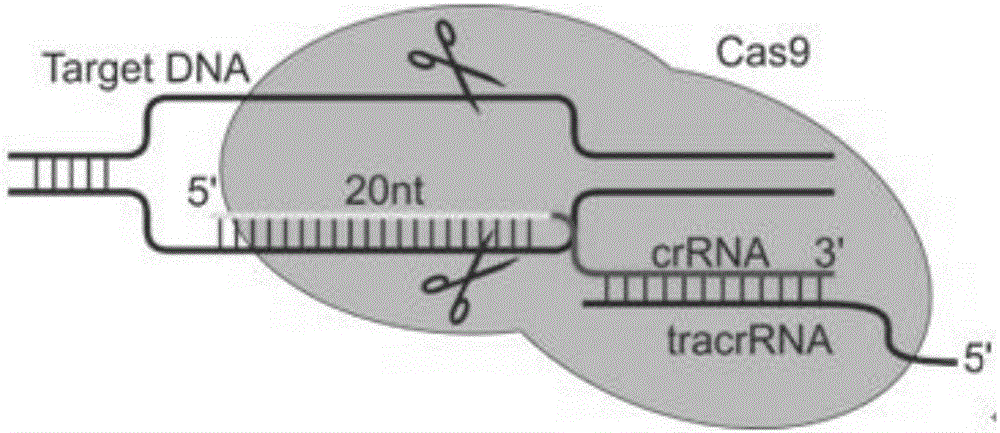
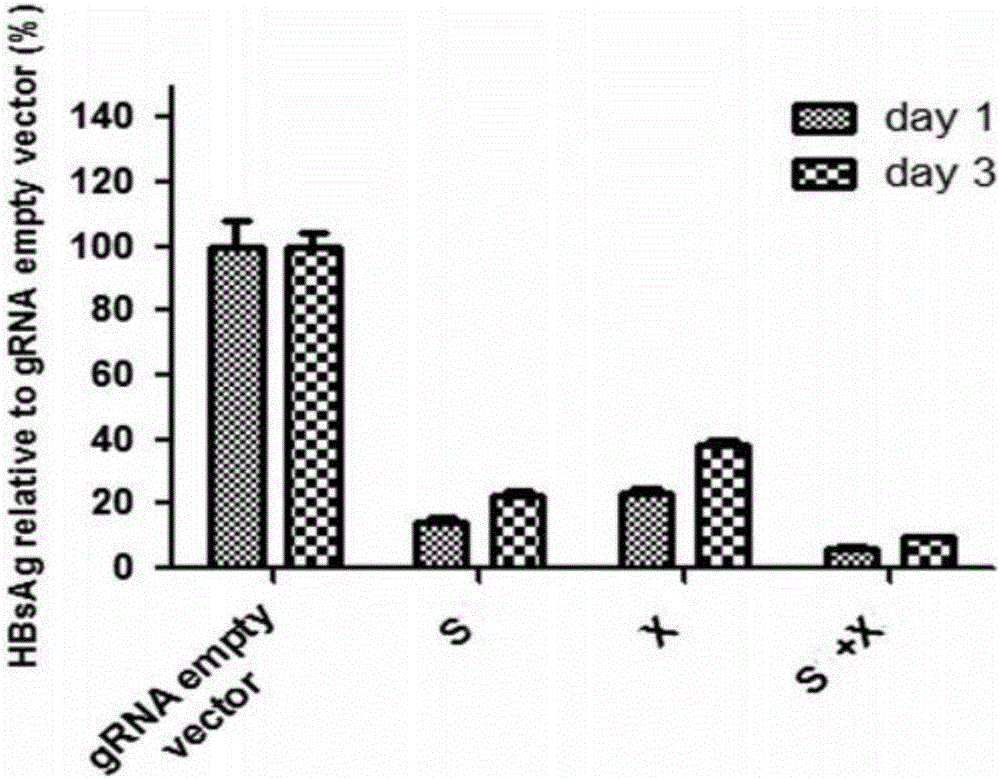
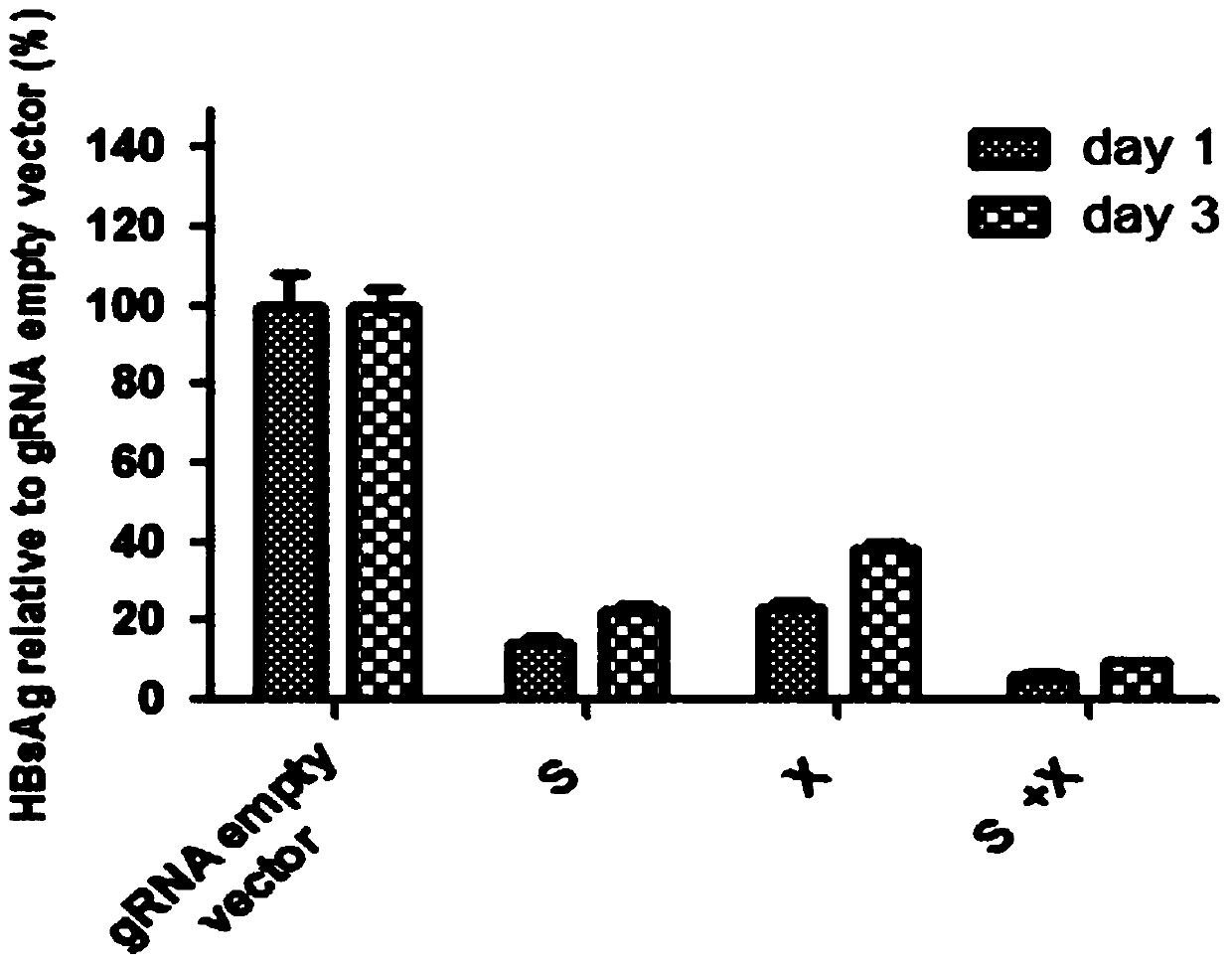
Specific sgRNA combined with immunogene to inhibit HBV replication, expression vector thereof, and application of specific sgRNA and expression vector
ActiveCN105821039AImprove targetingHigh knockout efficiencyOrganic active ingredientsGenetic material ingredientsHepatitis B Virus AntigenIn vivo
The invention provides a specific sgRNA combined with an immunogene to inhibit HBV replication, an expression vector thereof, and an application of the specific sgRNA and the expression vector. The sgRNA sequences of a human hepatitis B virogene suitable for CRISPR-Cas9 targeting editing and a PD-1 gene are designed, the plasmid vector of the sgRNA inhibiting HBV and PD-1 genes is constructed, and the plasmid vector and a nuclease gene expression vector are transferred to HBV transgenic mice, and can obviously inhibit HBV DNA replication. The gene expression vector prepared in the invention has the advantages of simple method steps, good sgRNA targeting property, and high CRISPR-Cas9 knockout efficiency. The sgRNAs specifically targeting the HBV and PD-1 genes can accurately splice the HBV and PD-1 genes, inhibit in vivo hepatitis B virus replication, and reduce the hepatitis B virus antigen expression.
Owner:李旭
Method for pig CMAH gene specific knockout through CRISPR-Cas9 and sgRNA for specially targeting CMAH gene
The invention discloses a method for pig CMAH gene specific knockout through CRISPR-Cas9 and sgRNA for specially targeting a CMAH gene. A target sequence of the sgRNA for specially targeting the CMAH gene on the CMAH gene meets a 5'-N(20)NGG-3' sequence arrangement rule, wherein the N(20) represents 20 continuous bases, and each N represents A, T, C or G; the target sequence on the CMAH gene locates at a 5-exon coding area or a junction position of adjacent introns of an N end of the CMAH gene; and the target sequence on the CMAH gene is unique. The sgRNA is used in a method for pig CMAH gene specific knockout through the CRISPR-Cas9, the pig CMAH gene can be rapidly, accurately, efficiently and specifically knocked out, and the problems of the long period and high cost of construction of a CMAH gene knockout pig are effectively solved.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Method for CRISPR-Cas9 specific knockout of pig GGTA1 gene and sgRNA for specific targeted GGTA1 gene
The invention discloses a method for CRISPR-Cas9 specific knockout of a pig GGTA1 gene and sgRNA for a specific targeted GGTA1 gene. The target sequence of the sgRNA for the specific targeted GGTA1 gene on the GGTA1 gene is in accordance with the sequence arrangement rule of 5'-N(20)NGG-3', wherein N(20) represents 20 continuous bases, and each N represents A or T ot C or G; the target sequence on the GGTA1 gene is positioned at a junction of 5 exon coding areas and / or adjacent introns of the N-end of the GGTA1 gene; and the target sequence on the GGTA1 gene is unique. According to the method for CRISPR-Cas9 specific knockout of the pig GGTA1 gene via sgRNA, specific knockout of the pig GGTA1 gene can be rapidly and accurately realized with high efficiency, and the problems of long period and high cost for constructing the knockout of the pig GGTA1 gene can be effectively solved.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Monocotyledon plant gene knockout vector based on CRISPR/Cas9 technology, and applications thereof
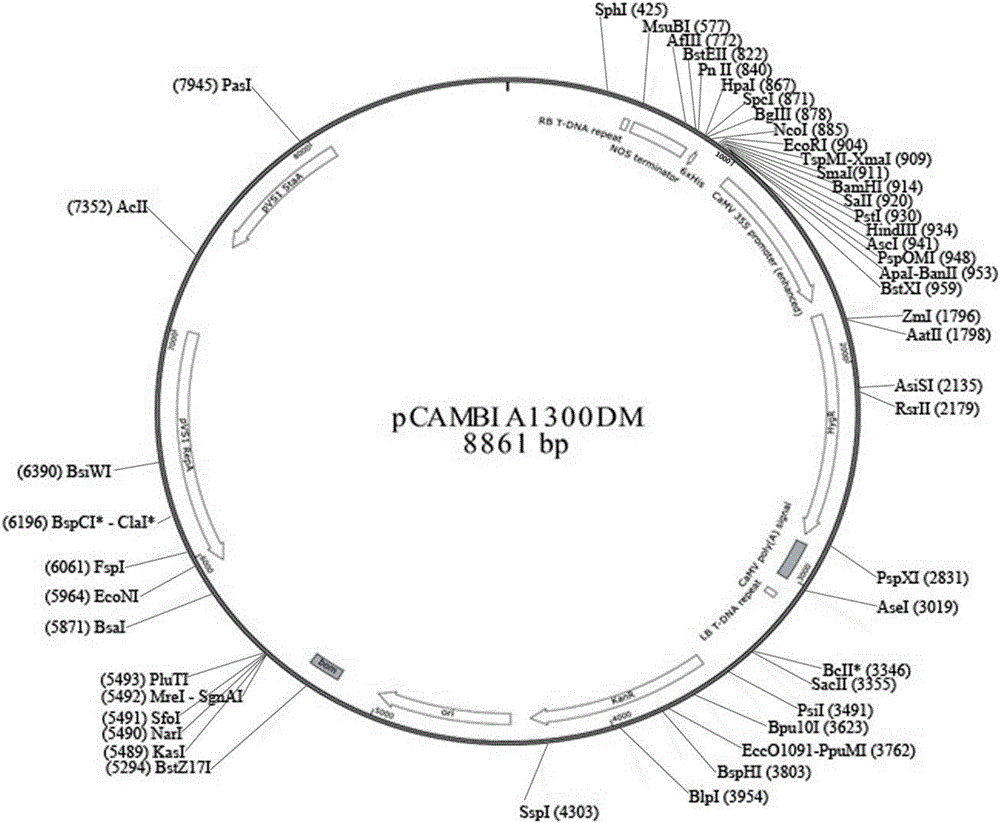
InactiveCN106167810AEfficient constructionKnockout EfficientNucleic acid vectorPlant peptidesDigestionInsertion site
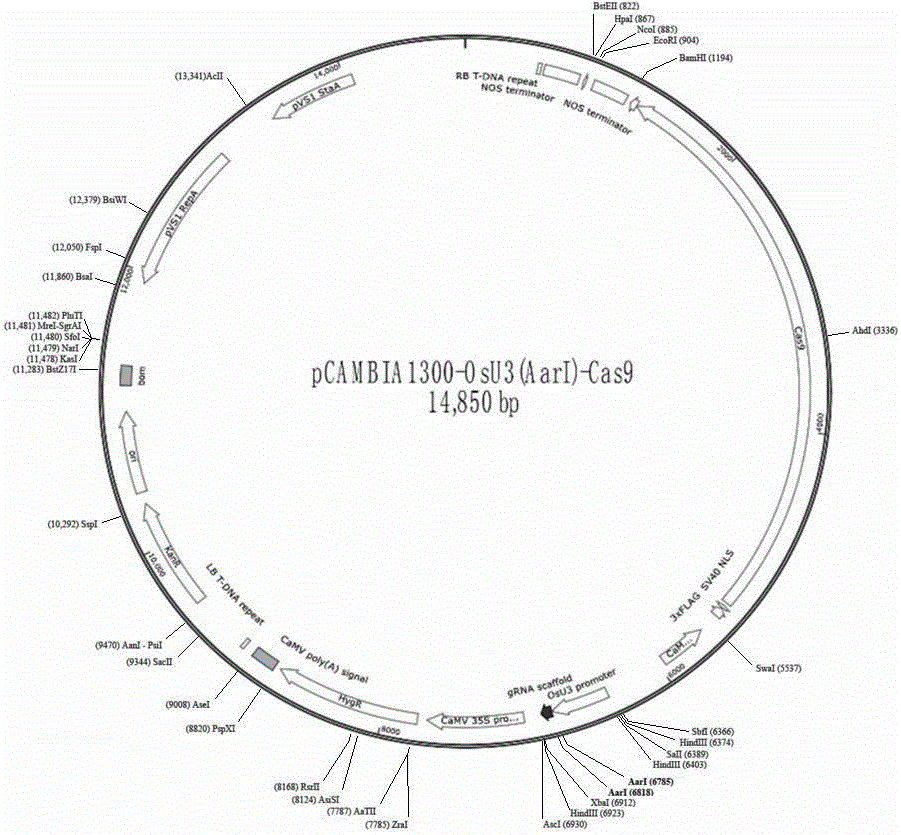
The invention discloses a plant gene knockout vector based on a CRISPR / Cas9 technology. The monocotyledon plant gene knockout vector is characterized in that the plant gene knockout vector is a monocotyledon plant CRISPR / Cas9 knockout vector pCambin1300-OsU3-Cas9, and contains the AarI insertion site. According to the present invention, with the CRISPR / Cas9 plant knockout double-component vector pCambin1300-OsU3-2x35s-Cas9 formed from a sgRNA-OsU3 structure sequence comprising the AarI insertion site and a 2x35s-Cas9-ter sequence, the purpose of the efficient and rapid construction of the monocotyledon plant gene knockout vector can be achieved through the one-step digestion linkage method; and the provided CRISPR / Cas9 knockout vector pCambin1300-OsU3-2x35s-Cas9 has characteristics of high targeting efficiency and extremely low undershoot efficiency, and can be used for plant target gene knockout or can be used for plant gene editing in a kit form.
Owner:内蒙古中科正标生物科技有限责任公司
Method using CRISPR-Cas9 to specifically knock off pig PDX1 gene and sgRNA of PDX1 gene for specific targeting
The invention discloses a method using CRISPR-Cas9 to specifically knock off pig PDX1 gene and sgRNA of PDX1 gene for specific targeting. The target sequence of sgRNA of PDX1 gene for specific targeting on PDX1 gene is accord with the sequence alignment rule of 5'-N(20)NGG-3', wherein N(20) represents 20 continuous bases, and each N represent A, T, C or G; the target sequence of PDX1 gene is arranged in the first exon coding region on the N terminal of PDX1 gene or a junction between the coding region and neighbored intron, and the target sequence of PDX1 gene is unique. The provided method can rapidly, precisely, efficiently and specifically knock off PDX1 gene of pigs, and solves the problems of long period and high cost of pig PDX1 gene knocking.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
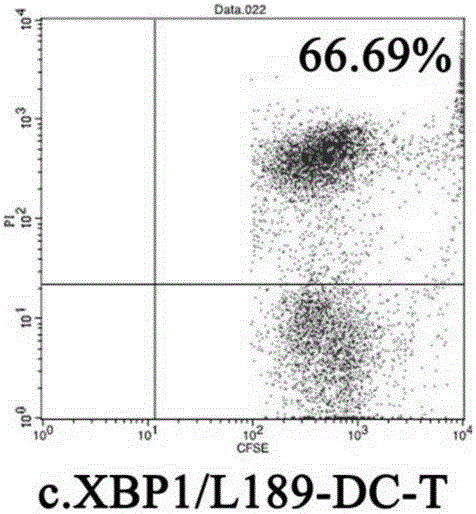
High-efficiency knockout method for XBP1 gene in DC cell
InactiveCN105602987AGuaranteed immune stimulationEffective anti-tumor immunityBlood/immune system cellsVector-based foreign material introductionCompetent cellXBP1
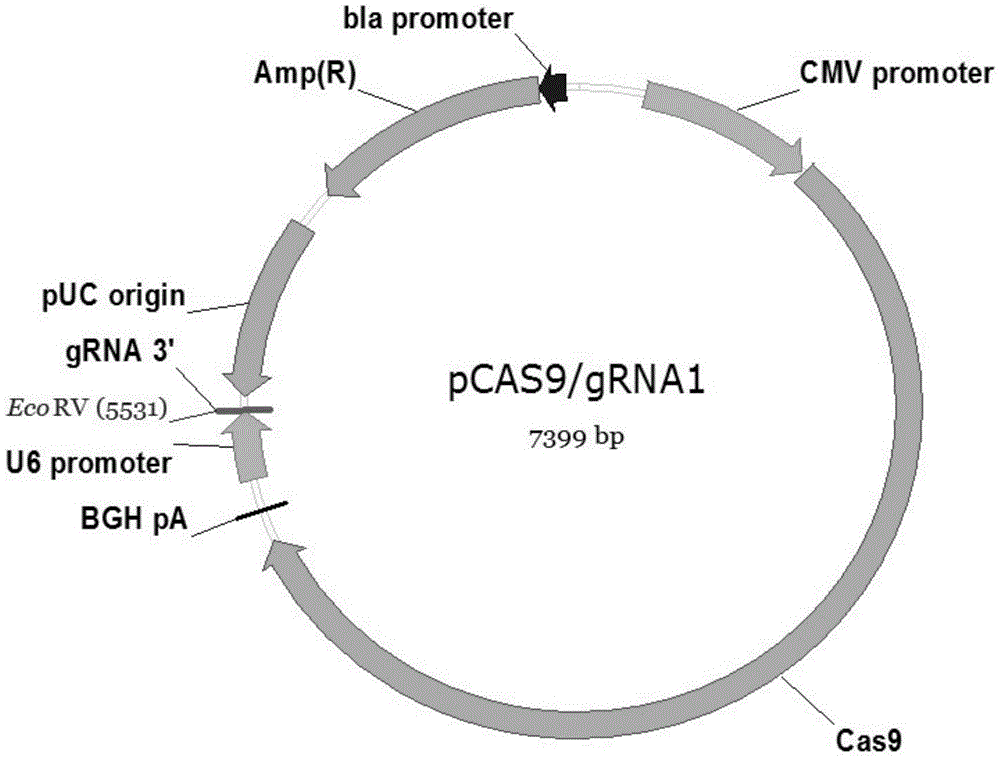
The invention provides a high-efficiency knockout method for the XBP1 gene in a DC cell. The high-efficiency knockout method for the XBP1 gene in the DC cell comprises the following steps: designing of a gene knockout target and oligonucleotide; annealing of oligonucleotide; enzyme digestion of a linearized vector; connection and reaction of the linearized vector with double-strand oligonucleotide; transformation of competent cells and knockout of the XBP1 gene in the DC cell; etc. According to the invention, the knockout method provided by the invention employs an improved CRISPR / pCas9 gene knockout system, uses the XBP1 as a target gene for designing of a CRISPR targeting sequence and preparation of pCas9 / gRNA1-XBP1 plasmid and further allows the plasmid to transfect the DC cell under the treatment action of an L189 drug; thus, the XBP1 gene in the DC cell can be effectively knocked out, the immunostimulation effect of the DC cells can be effectively guaranteed, and antineoplastic immunization effect of the DC cell is given to effective play.
Owner:SHENZHEN MORECELL BIOMEDICAL TECH DEV CO LTD
CRISPR/Cas9 technology-based monocotyledon gene knockout vector and application thereof
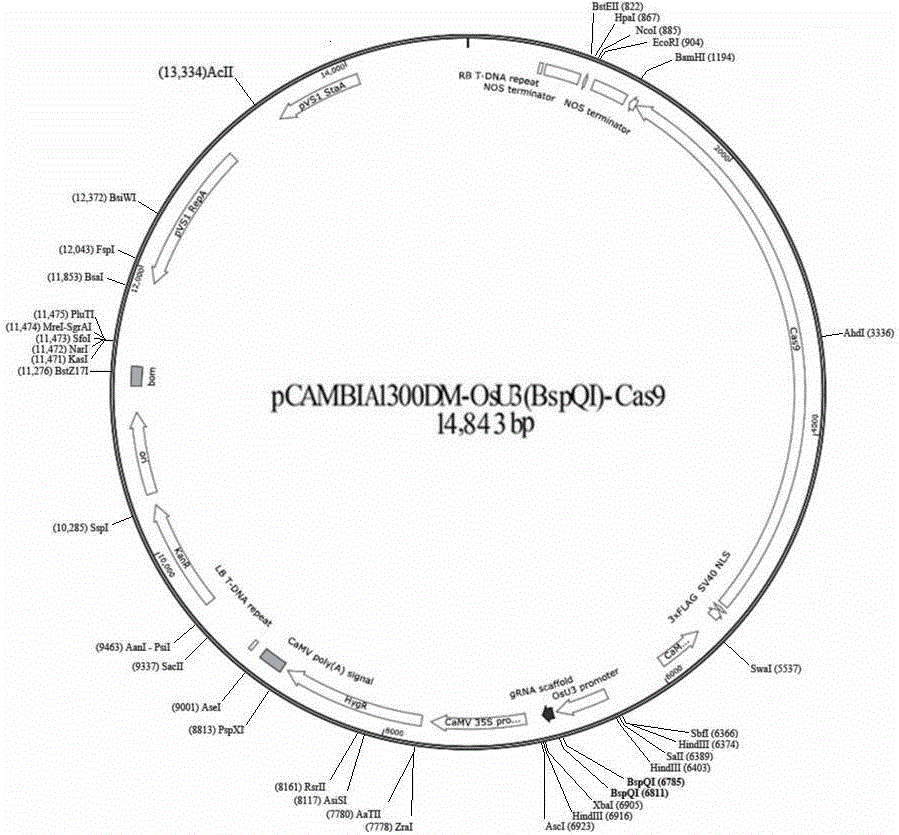
InactiveCN106434737AKnockout EfficientKnockout fastNucleic acid vectorVector-based foreign material introductionInsertion siteOff targets
The invention discloses a CRISPR / Cas9 technology-based plant gene knockout vector and an application thereof. The plant gene knockout vector is monocotyledon CRISPR / Cas9 knockout vector pCAMBIA1300DM-OsU3(BspQ I)-Cas9, and contains a BspQ I insertion site. The CRISPR / Cas9 plant knockout binary vector pCAMBIA1300DM-OsU3(BspQ I)-Cas9 commonly constructed by an sgRNA-OsU3 sequence composed of the BspQ I insertion site and a 2x35s-hSpCas9-ter sequence can realize high-efficiency rapid construction of the monocotyledon gene knockout vector through a one-step restriction and ligation method; and the CRISPR / Cas9 knockout vector pCAMBIA1300DM-OsU3(BspQ I)-Cas9 has the characteristics of high targeting efficiency and low off-target efficiency, and can be applied to plant target gene knockout or applied to plant gene editing in a kit form.
Owner:内蒙古中科正标生物科技有限责任公司
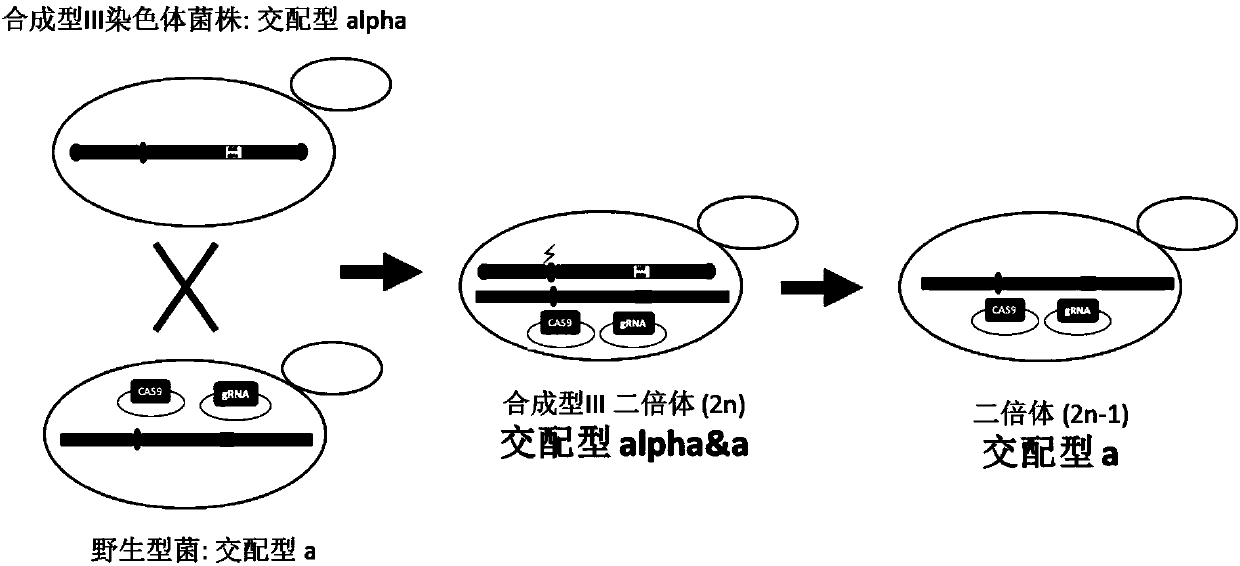
Method for knocking down brewing yeast chromosome
ActiveCN107858346AKnockout simpleKnockout EfficientHydrolasesStable introduction of DNABiotechnologyYeast chromosome
The invention relates to the technical field of biology, and specifically discloses a method for knocking down brewing yeast chromosome. The method is characterized in that the complete yeast chromosome is cut according to the CRISPR / Cas9 base technology; a specificity target close to centromere is selected; a corresponding guide RNA is designed to guide Cas9 protein to generate an incision closeto the centromere, thus realizing the knocking down of the whole chromosome. Compared with the conventional method for losing the whole chromosome in manners such as inducing through a Ga1 promoter, and reductional division, the method has the advantages that the brewing yeast chromosome is simply, efficiently and quickly cut; the cross exchange of sister chromatids can be avoided; the chromosome-knocked-down homozygous brewing yeast diploid strain can be obtained.
Owner:TIANJIN UNIV
sgRNA sequence for specifically targeting arabidopsis ILK2 gene and application of sgRNA sequence
InactiveCN107236737AKnockout EfficientKnockout fastHydrolasesMicrobiological testing/measurementNucleotide sequencingArabidopsis thaliana
The invention discloses a sgRNA sequence for specifically targeting an arabidopsis ILK2 gene and application of the sgRNA sequence. The nucleotide sequence of the sgRNA sequence is represented by SEQ ID NO.1. Two single-chain oligo DNA sequences are designed and synthesized according to a sgRNA guide sequence, a double chain is formed through annealing and is linked with a Cas9 carrier, a sgRNA coding sequence and a CRISPR system are introduced into Arabidopsis by using an agrobacterium-mediated genetic transformation technique, a target sequence is shorn by a Cas9 protein under the guidance of sgRNA, and therefore, the knockout of an ILK2 gene is realized. According to the sgRNA sequence provided by the invention, the ILK2 gene can be knocked out or edited by virtue of a CRISPR-Cas9 system, so as to analyze the functions of the arabidopsis ILK2 gene.
Owner:SHANGHAI JIAO TONG UNIV
Establishment of carrier based on fish CRISPR/Cas9 system by using gene knockout method ad establishing method of carrier
InactiveCN104232669AEasy to operateHigh targeting accuracyVector-based foreign material introductionTransgenesisElectroporation
The invention discloses establishment of a carrier based on a fish CRISPR / Cas9 system by using a gene knockout method ad an establishing method of the carrier, and relates to a carrier based on the fish CRISPR / Cas9 system and an establishing method of the carrier, so as to solve the problems that in a conventional transgenosis method the position that transgenosis is inserted into genome is hard to confirm and potential safety hazard can be generated when external DNA is led into cells and is randomly integrated by using a microscopic injection method, an electroporation method, a sperm carrier method and the like, and the problems that a conventional gene knockout method is complex in process, high in requirement on techniques, high in expense and long in experiment period, and the success rate is limited by multiple factors. By co-ejecting two non-integrated knockout carriers, namely, pX330-MSTN a and pX330-MSTN b, into zygotes of fishes in the 1-2 cell stages, high-efficiency and rapid gene knockout can be achieved.
Owner:HEILONGJIANG RIVER FISHERY RES INST CHINESE ACADEMY OF FISHERIES SCI
Tumor-targeting gene therapy drug based on CRISPR/Cas9 gene editing technology and use thereof
PendingCN109971755ARapid and efficient knockoutEasy to inactivateOrganic active ingredientsHydrolasesDNMT1 GeneTumor targeting
The present invention provides an sgRNA specifically targeting a human DNMT1 gene based on a CRISPR / Cas9 system, a use thereof for guiding the CRISPR / Cas9 system in a drug for treatment and / or prevention of tumors, and a method correspondingly constructing a sgRNA expression vector. The CRISPR / Cas9 system can rapidly, efficiently and specifically knock out the human DNMT1 gene, induces inactivation of the DNMT1 gene in tumor cells, inhibits expression of the DNMT1 and methylation of DNA and thus inhibits tumor cell growth.
Owner:CHENGDU JINKAI BIOTECH CO LTD +1
Method for quickly building CRISPR gene editing liver cancer cell strain and cell strain
PendingCN111254164AGood expression efficiencyStable knockoutHydrolasesGenetically modified cellsCas9Liver adenocarcinoma
The invention discloses a method for quickly building a CRISPR gene editing liver cancer cell strain. According to the method disclosed by the invention, a CRISPR / Cas 9 technique is improved, recombinant plasmids better in expression efficiency are constructed, quick monoclone culture is combined, and a stable gene knockout liver cancer cell strain is constructed. Required sgRNA is accurately obtained through primer synthesis, an inserting fragment is synthetized through two-step PCR, a carrier is loaded, and recombinant plasmids for knockout are constructed; after slow viruses are packaged, the packaged slow viruses and liver cancer cells are co-incubated, and sgRNA and Cas9 proteins of equal quantity are transmitted into the liver cancer cells at the same time through slow virus mediating; and liver cancer cells after gene editing are subjected to puromycin resistance screening and monoclone culture, and finally, a gene knock-out positive stable liver cancer cell strain is quickly obtained. According to the method disclosed by the invention, important experimental materials are provided for researching a molecular mechanism of the gene in generation and development of tumors, andreference is provided for in vitro cell modeling of liver cancer diseases.
Owner:DALIAN INST OF CHEM PHYSICS CHINESE ACAD OF SCI
Kit for knocking out glioblastoma DHC2 gene
ActiveCN107841509AKnockout EfficientKnockout stableStable introduction of DNAAnimals/human peptidesGlioblastoma cellAllele
The invention relates to a kit for knocking out a glioblastoma DHC2 gene, and application of the kit, and a method for knocking out a glioblastoma DHC2 allele. The kit for knocking out the glioblastoma DHC2 gene comprises a first knockout plasmid and a second knockout plasmid, or a knockout plasmid and a third knockout plasmid, or comprises a first knockout plasmid, a second knockout plasmid and athird knockout plasmid, wherein a first guide RNA shown in a sequence SEQ ID NO: 1 is transcribed from the first knockout plasmid, and a second guide RNA shown in a sequence SEQ ID NO: 2 is transcribed from the second knockout plasmid; a third guide RNA shown in a sequence SEQ ID NO: 3 is transcribed from the third knockout plasmid. The kit for stably knocking out the DHC2 gene is firstly provided. The knockout plasmid included in the kit has the characteristics of low off-target rate and strong specificity. The kit can stably knock out two DHC2 alleles, and has extremely high gene editing efficiency.
Owner:NANFANG HOSPITAL OF SOUTHERN MEDICAL UNIV
Method for specifically knocking out fumarylacetoacetate hydrolase (FAH) gene by using CRISPR-Cas9 and specific sgRNA
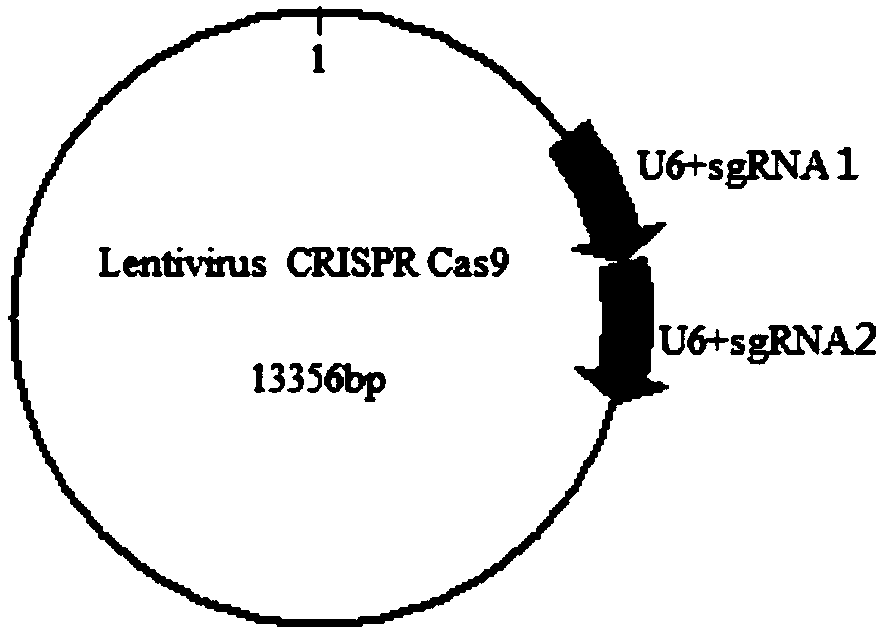
PendingCN111100876AKnockout fastKnockout precisionHydrolasesStable introduction of DNABiotechnologyLentivirus
The invention relates to a method for specifically knocking out a fumarylacetoacetate hydrolase (FAH) gene by using CRISPR-Cas9 and specific sgRNA. The method is applied to specifically knock out theFAH gene of pigs and rabbits and comprises the following specific steps: S1: selection and design of a sgRNA target sequence; S2: construction of a CRISPR-Cas9 vector of the FAH gene; S3: obtaining ofa pseudotype lentivirus expressing FAH sgRNA and Cas9 protein; and S4: infection of a target cell and detection of an FAH gene knockout effect. The target sequence of the specific sgRNA on the FAH genes of the pigs and rabbits conforms to a sequence arrangement law of 5'-N(20)NGG-3'. The specific sgRNA is applied to the method for specifically knocking out the FAH gene of the pigs and rabbits byusing the CRISPR-Cas9, can rapidly, accurately, efficiently and specifically knock out the FAH gene of the pigs and rabbits respectively, and effectively solves the technical problems of long period and high cost in construction of pigs and rabbits with the FAH gene knocked out.
Owner:立沃生物科技(深圳)有限公司
CRISPR/Cas9 vector suitable for paraconiothyrium hawaiiense FS482 as well as construction method and application of CRISPR/Cas9 vector
ActiveCN112553238AHigh knockout efficiencyKnockout EfficientMicroorganism based processesEnzymesBiosynthetic genesSecondary metabolite
The invention discloses a CRISPR / Cas9 vector suitable for paraconiothyrium hawaiiense FS482 as well as a construction method and application of the CRISPR / Cas9 vector. According to the invention, a recombinant paraconiothyrium hawaiiense FS482 strain with knocked-out diterpenoid new skeleton compound biosynthetic genes is constructed by utilizing a novel CRISPR / Cas9 system after promoter optimization for the first time, and an efficient CRISPR / Cas9 gene knockout system suitable for deep sea fungus paraconiothyrium hawaiiense FS482 is established; and therefore, a molecular biological foundation is laid for clarification of a biosynthesis mechanism of the novel active secondary metabolite of the paraconiothyrium hawaiiense FS482 and obtaining of more novel secondary metabolites with remarkable biological activity.
Owner:GUANGDONG INST OF MICROBIOLOGY GUANGDONG DETECTION CENT OF MICROBIOLOGY
Cell line for targeted knockout of pig GDPD2 gene based on CRISPR-Cas9 technology, and construction method of cell line
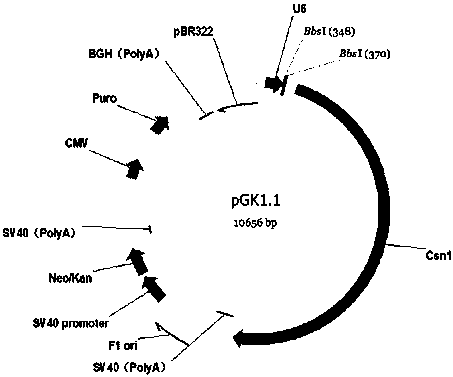
PendingCN112011539AContribute to researchContributes to protein functionHydrolasesGastrointestinal cellsNegative strandDouble strand
The invention discloses a cell line for the targeted knockout of a pig GDPD2 gene based on the CRISPR-Cas9 technology, and a construction method of the cell line. The cell line is prepared by the following steps: (1) designing an sgRNA guide sequence according to the pig GDPD2 gene; (2) annealing a positive strand sgRNA sequence and a negative strand sgRNA sequence to form double-stranded DNA, andconnecting the double-stranded DNA with a linearization pGK1.1 vector to obtain a positive targeting vector; and (3) performing mixed electrotransfection on the positive targeting vector and target cells IPEC-J2 to obtain GDPD2 gene knockout IPEC-J2 cells. The CRISPR / Cas9 technology is used for establishing the pig GDPD2 gene knockout IPEC-J2 cell line for the first time, and the research on theprotein function of GDPD2 is facilitated. The CRISPR / Cas9 knockout technology is simple in method, the target gene can be efficiently knocked out by designing sgRNA to cause that the function of the GDPD2 gene is lost, and thereby the pig GDPD2 gene knockout IPEC-J2 cell line is an ideal GDPD2 gene knockout IPEC-J2 cell model.
Owner:YANGZHOU UNIV
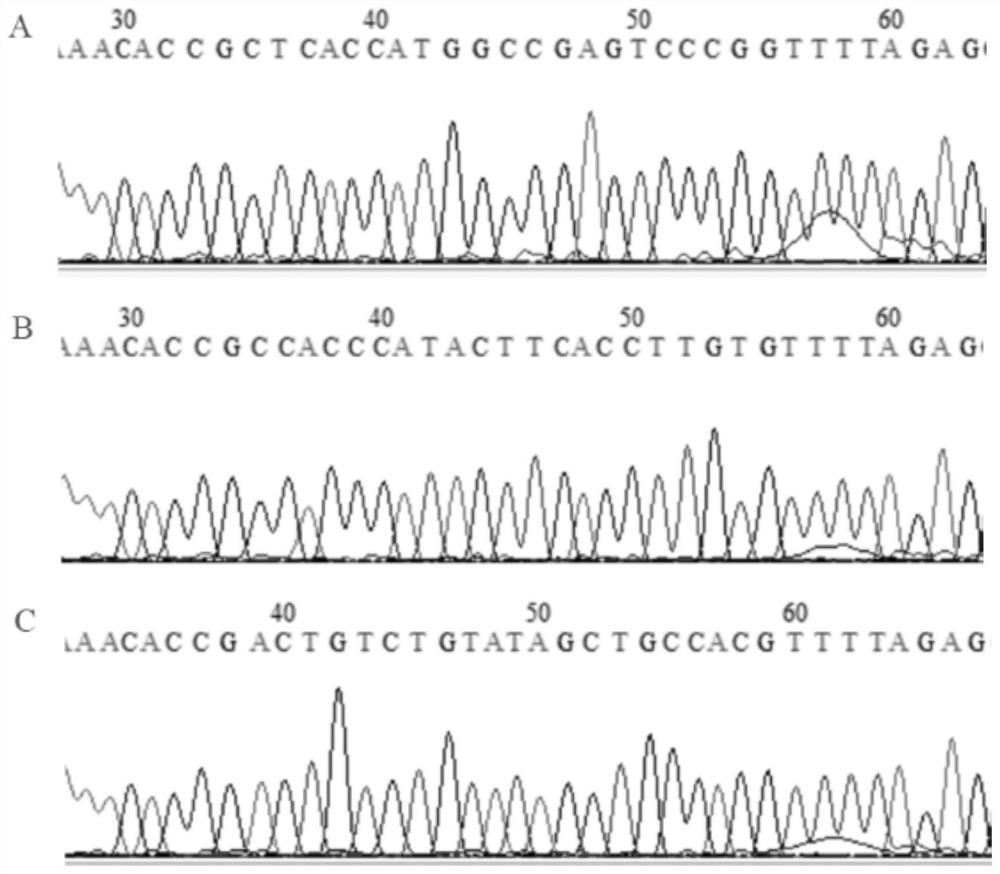
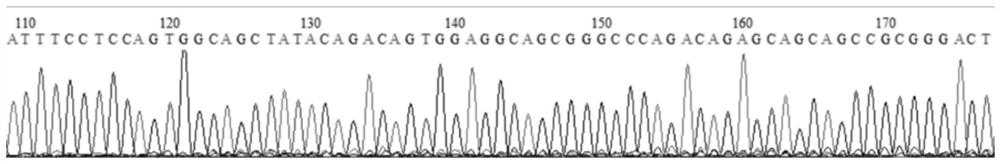
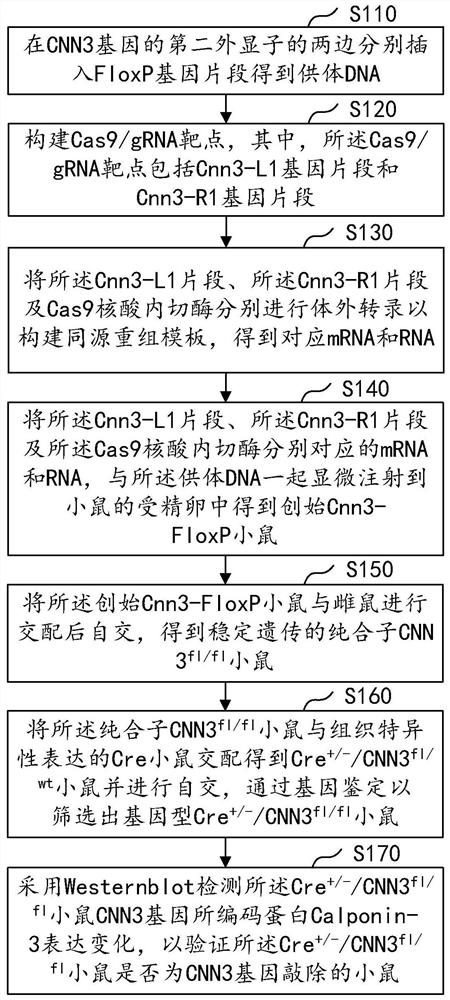
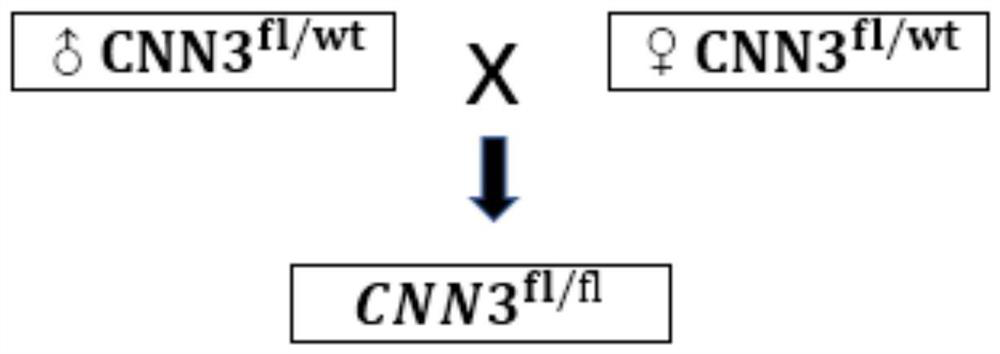
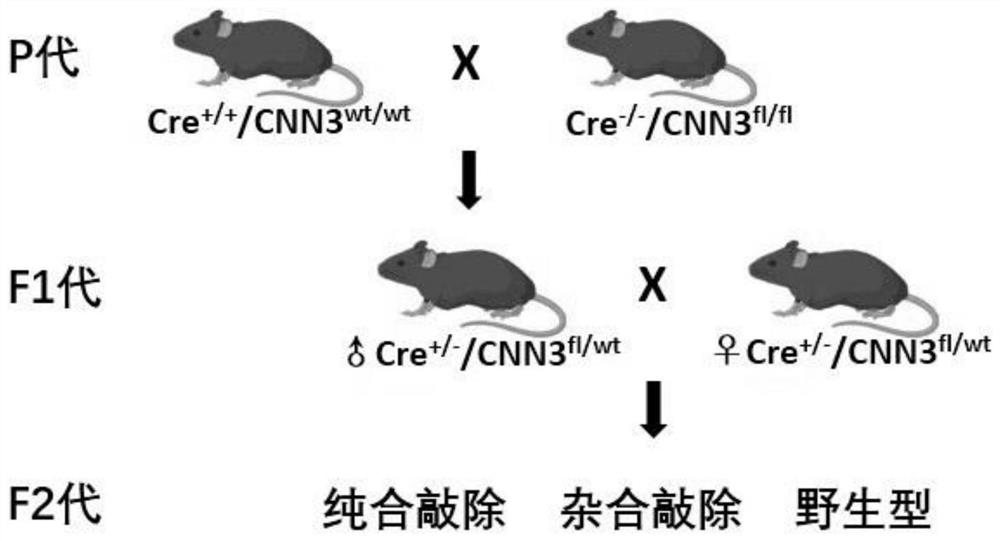
Construction method of CNN3 gene knockout mouse model based on Cre-FloxP system
PendingCN113584030AGenetic stabilityGenetically efficientHydrolasesStable introduction of DNAGenotypeGene recognition
The invention discloses a construction method of a CNN3 gene knockout mouse model based on a Cre-FloxP system, which comprises the following steps: respectively inserting FloxP gene segments at two sides of a second exon of a CNN3 gene, constructing a Cas9 / gRNA target and a homologous recombination template, carrying out microinjection on the homologous recombination template and donor DNA into a fertilized egg of a mouse to obtain a Cnn3-FloxP mouse, and carrying out Cre-FloxP system-based CNN3 gene knockout mouse model; after mating with a female mouse, conducting selfing to obtain a stably inherited homozygote CNN3fl / fl mouse; after the homozygote CNN3fl / fl mouse and a Cre mouse with tissue specific expression are copulated, conducting selfing, and screening out the mouse with the genotype of Cre + / - / CNN3fl / fl through gene recognition. The invention relates to the technical field of gene design, can efficiently construct a mouse with specific tissue or specific cell CNN3 gene knockout, and greatly improves the construction efficiency and the construction success rate of the mouse model with the selective CNN3 knockout function.
Owner:FIRST AFFILIATED HOSPITAL OF KUNMING MEDICAL UNIV
CRISPR-Cas9-specific method for knocking out porcine SALL1 gene and sgRNA for specifically targeting SALL1 gene
ActiveCN105518137BKnockout fastKnockout precisionFermentationVector-based foreign material introductionBase JGenetics
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Crispr/cas9 system to knock out the pds gene of Populus alba and its application
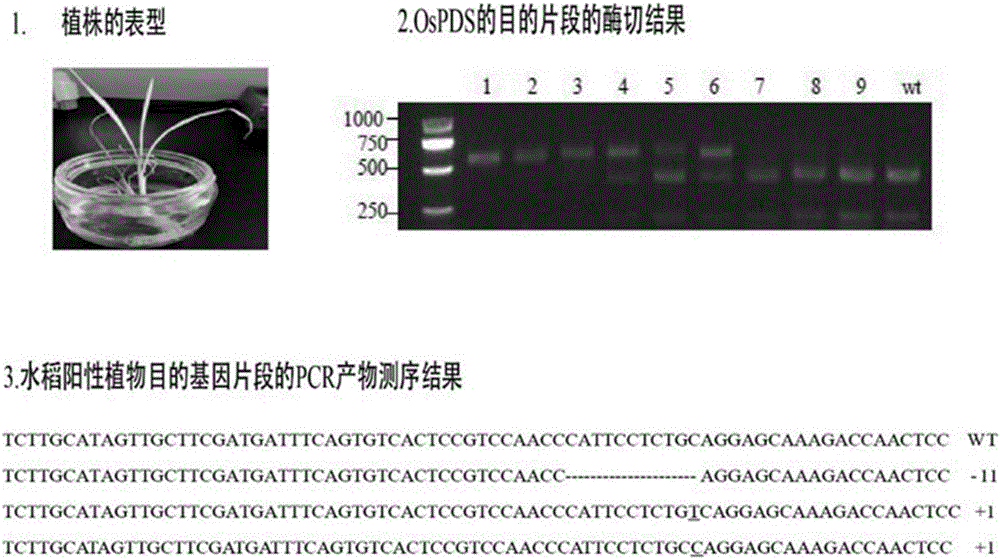
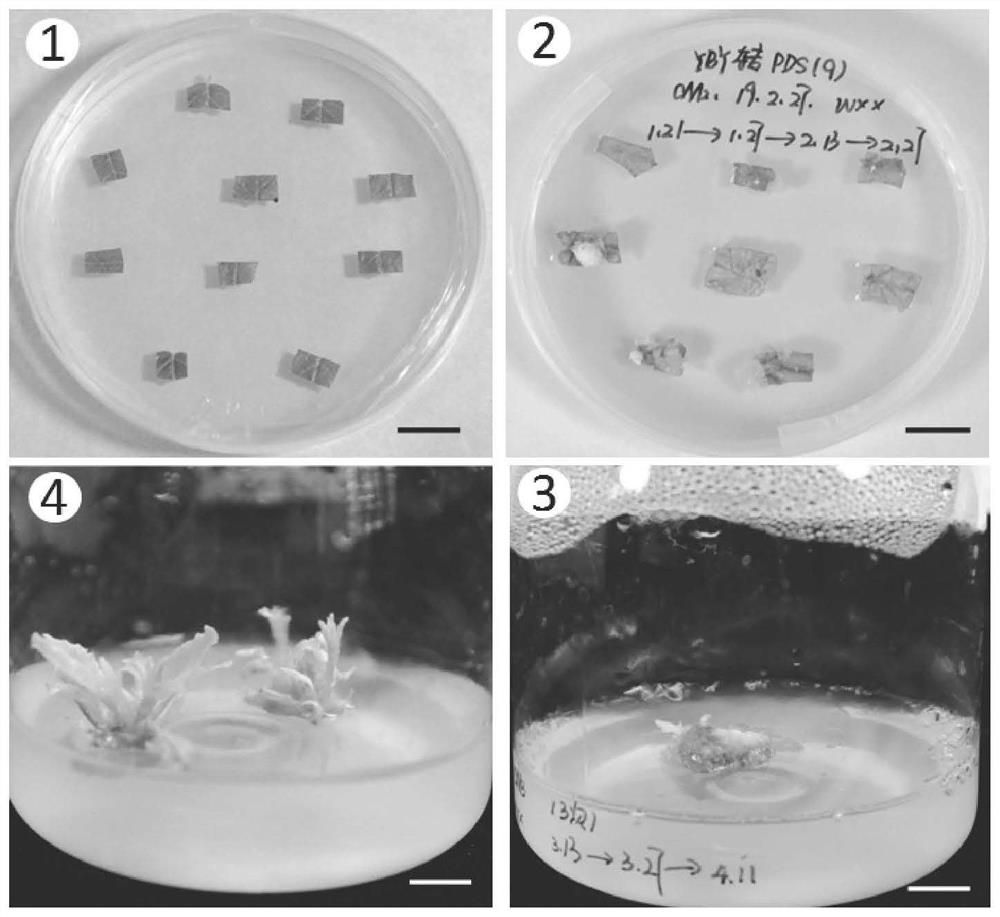
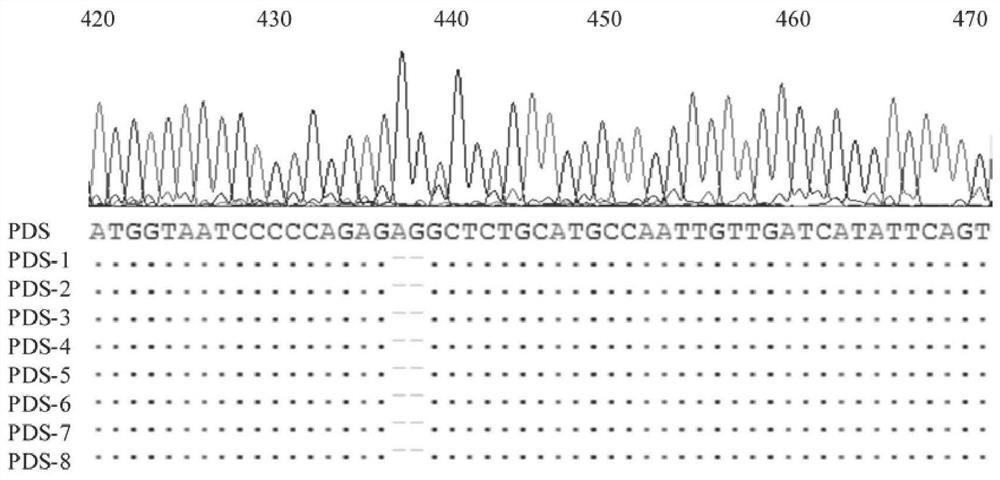
ActiveCN111235177BFacilitate the study of gene functionKnockout EfficientVector-based foreign material introductionAngiosperms/flowering plantsBiotechnologyLycopersene
The invention relates to a CRISPR / Cas9 system knockout PDS gene of Populus alba and its application, and belongs to the technical field of molecular breeding. The nucleotide sequence of the Populus alba phytoene dehydrogenase (PDS) gene of the present invention is shown in SEQ ID NO.1. The target site described in the present invention can realize site-directed editing of the PDS gene, and the nucleotide sequence of the target site is shown in SEQ ID NO.2. The invention provides a new scheme for gene function research of woody model plants, establishes a molecular breeding platform for native tree species of my country's poplar, and provides technical support for the directional cultivation of new forest species that meet the needs of my country's economic development.
Owner:CHINESE ACAD OF FORESTRY
A kind of sgRNA sequence and its application of specific knockout dihydrofolate reductase gene
ActiveCN107201365BKnockout fastKnockout EfficientNucleic acid vectorOxidoreductasesForeign proteinDihydrofolic acid
The invention provides a sgRNA sequence for specifically knocking-out a DHFR (Dihydrofolate Reductase) gene. The sequence is as shown in SEQ ID NO:3. The invention also provides the application of the sgRNA sequence and a kit for knocking-out the DHFR gene. The sgRNA which is provided by the invention can be used for knocking-out the DHFR gene in a CHO (Chinese Hamster Ovary) cell quickly, efficiency and specifically, and obtaining a DHFR gene-deleted cell line; the yield of foreign protein can be greatly improved; the application prospect is good.
Owner:四川丰讯科技发展有限公司
CRISPR-Cas9-specific method for knocking out porcine CMah gene and sgRNA for specifically targeting CMah gene
The invention discloses a method for pig CMAH gene specific knockout through CRISPR-Cas9 and sgRNA for specially targeting a CMAH gene. A target sequence of the sgRNA for specially targeting the CMAH gene on the CMAH gene meets a 5'-N(20)NGG-3' sequence arrangement rule, wherein the N(20) represents 20 continuous bases, and each N represents A, T, C or G; the target sequence on the CMAH gene locates at a 5-exon coding area or a junction position of adjacent introns of an N end of the CMAH gene; and the target sequence on the CMAH gene is unique. The sgRNA is used in a method for pig CMAH gene specific knockout through the CRISPR-Cas9, the pig CMAH gene can be rapidly, accurately, efficiently and specifically knocked out, and the problems of the long period and high cost of construction of a CMAH gene knockout pig are effectively solved.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Specific sgRNA, expression vector and application of combined immune gene inhibition of HBV replication
ActiveCN105821039BImprove targetingHigh knockout efficiencyOrganic active ingredientsGenetic material ingredientsAntigenHepatitis B Virus Antigen
The invention provides a specific sgRNA combined with an immunogene to inhibit HBV replication, an expression vector thereof, and an application of the specific sgRNA and the expression vector. The sgRNA sequences of a human hepatitis B virogene suitable for CRISPR-Cas9 targeting editing and a PD-1 gene are designed, the plasmid vector of the sgRNA inhibiting HBV and PD-1 genes is constructed, and the plasmid vector and a nuclease gene expression vector are transferred to HBV transgenic mice, and can obviously inhibit HBV DNA replication. The gene expression vector prepared in the invention has the advantages of simple method steps, good sgRNA targeting property, and high CRISPR-Cas9 knockout efficiency. The sgRNAs specifically targeting the HBV and PD-1 genes can accurately splice the HBV and PD-1 genes, inhibit in vivo hepatitis B virus replication, and reduce the hepatitis B virus antigen expression.
Owner:李旭
A kind of Trichoderma harzianum gene knockout method
ActiveCN108148876BPromote growthShort experiment cycleFungiStable introduction of DNABiotechnologyEtioplasts
Owner:NANJING AGRICULTURAL UNIVERSITY
Construction method of Mia3 conditional gene knockout mouse model
ActiveCN114606270AMeet the needs of different research directionsKnockout EfficientCompounds screening/testingHydrolasesKnockout animalCell biology
The invention relates to a method for constructing a Mia3 gene conditional knockout mouse model by using a CRISPR (clustered regularly interspaced short palindromic repeats) / Cas9 (CRISPR associated protein 9) technology. The method comprises the following steps: 1, designing sgRNA (small guide ribonucleic acid) aiming at mouse Mia3 gene Exon2-Exon7, 2, obtaining the sgRNA by using an in-vitro transcription technology, and 3, co-injecting or co-electrically transferring a targeting vector, the sgRNA and Cas9 protein to a mouse fertilized egg. Compared with a traditional ES targeting mouse model, the Mia3 gene conditional knockout mouse model constructed for the first time has the characteristics of high efficiency, rapidness, simplicity, convenience, low cost and the like, and the expression of the gene in a specific tissue or specific time can be regulated through Cre.
Owner:广东药康生物科技有限公司
CRISPR-Cas9-specific method for knocking out porcine PDX1 gene and sgRNA for specifically targeting PDX1 gene
The invention discloses a method for specifically knocking out pig PDX1 gene by using CRISPR-Cas9 and sgRNA for specifically targeting PDX1 gene. The target sequence of the sgRNA specifically targeting the PDX1 gene of the present invention on the PDX1 gene conforms to the sequence arrangement rule of 5'-N(20)NGG-3', wherein N(20) represents 20 consecutive bases, wherein each N represents A or T or C or G; the target sequence on the PDX1 gene is located in the first exon coding region of the N-terminal of the PDX1 gene or at the junction with the adjacent intron; the target sequence on the PDX1 gene only one. The sgRNA of the present invention is used in the method of CRISPR-Cas9 to specifically knock out the porcine PDX1 gene, which can quickly, accurately, efficiently and specifically knock out the porcine PDX1 gene, effectively solving the long period and high cost of constructing PDX1 knockout pigs The problem.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
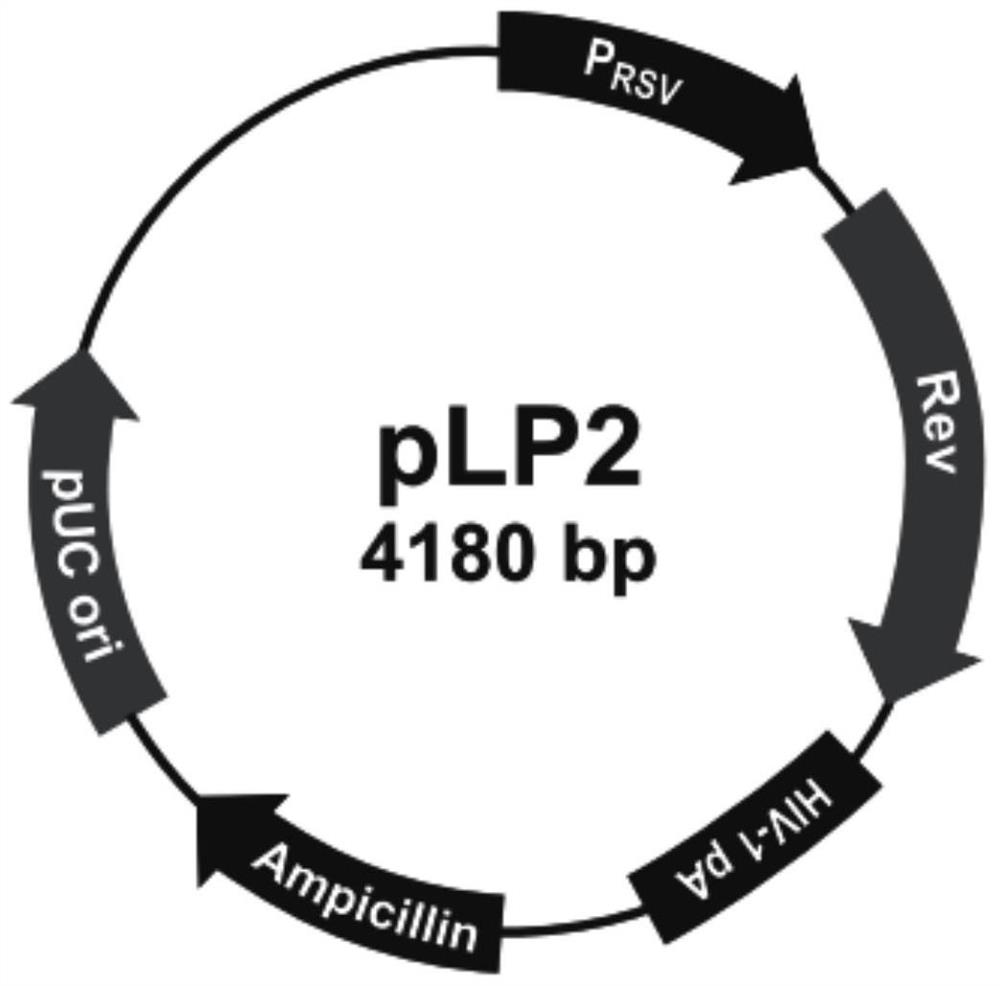
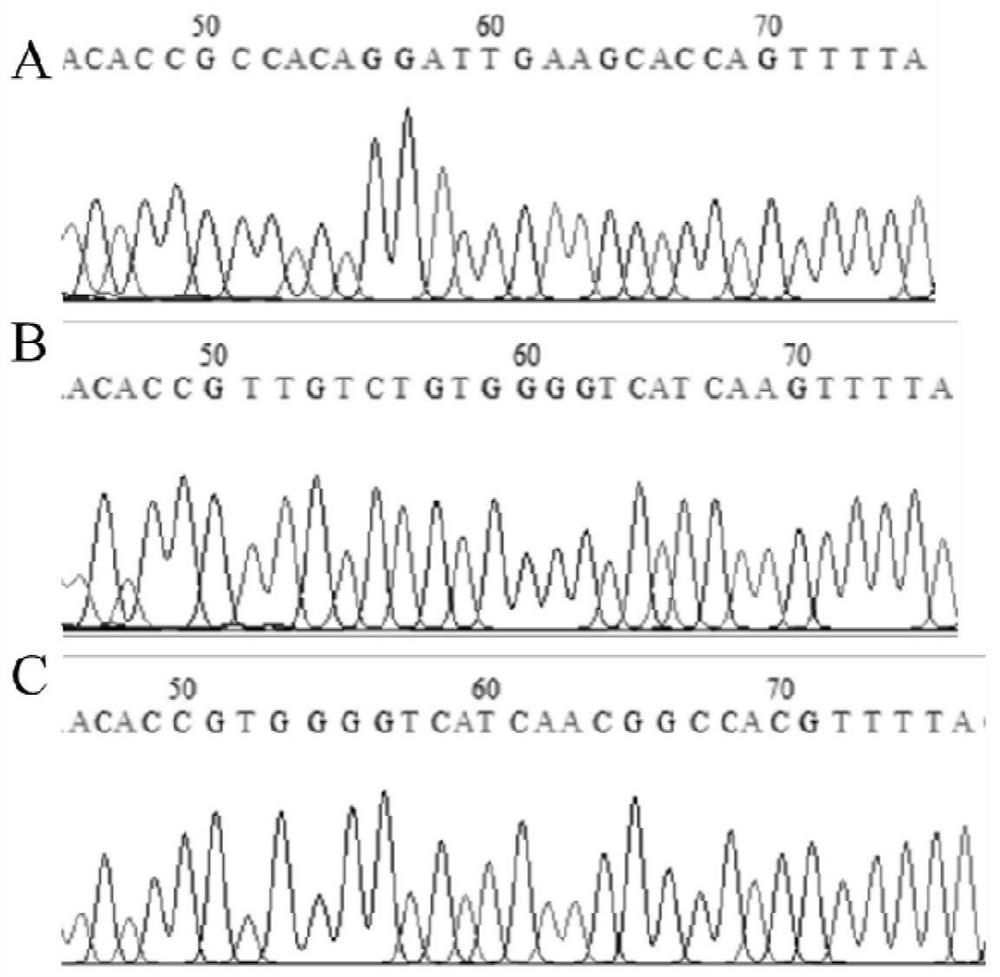
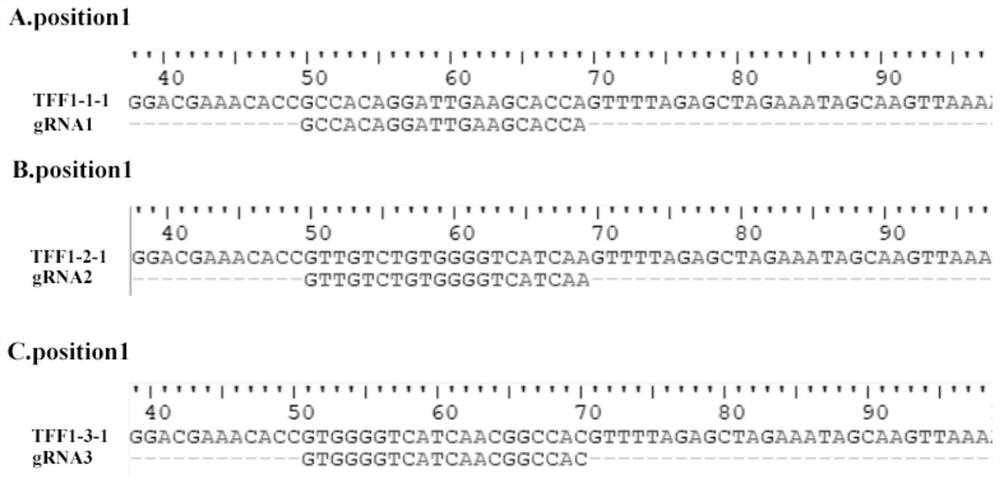
Construction method of porcine TFF1 gene knockout cell line based on CRISPR-Cas9 gene editing technology
PendingCN112921040AContribute to researchIdeal knockoutAnimals/human peptidesVector-based foreign material introductionGenome editingDisease resistant
The invention relates to a construction method of a porcine TFF1 gene knockout cell line based on a CRISPR-Cas9 gene editing technology. The TFF1 gene sequence in IPEC-J2 cells is knocked out by adopting the CRISPR / Cas9 technology, and the TFF1 gene knockout IPEC-J2 cell line is established and can be used for functional verification of a TFF1 gene in porcine breeding for disease resistance, and an action mechanism of the TFF1 gene in disease resistance is disclosed.
Owner:YANGZHOU UNIV
CRISPR/Cas9 systems and application of CRISPR/Cas9 systems in construction of alpha, beta and alpha-beta thalassemia model pig cell lines
ActiveCN112538497AImprove editing efficiencyKnockout EfficientHydrolasesGenetically modified cellsGenome editingFibroblastic cell
The invention discloses CRISPR / Cas9 systems and an application of the CRISPR / Cas9 systems in construction of alpha, beta and alpha-beta thalassemia model pig cell lines. A pair of target sequences isdesigned for each of pig HBA and HBB genes, three CRISPR / Cas9 systems are constructed by the two pairs of target sequences, each CRISPR / Cas9 system comprises a gRNA carrier and a Cas9 expression carrier, the expression carriers in the three CRISPR / Cas9 systems are respectively transferred into pig fibroblasts in proportion, and the alpha, beta and alpha-beta thalassemia model pig cell lines are obtained by screening. Single gene mutation and combined gene mutation are respectively caused to the pig HBA and HBB genes in the pig primary fibroblasts through a gene editing technology, and HBA, HBBand HBA-HBB gene mutant cells are obtained.
Owner:NANJING KGENE GENETIC ENG CO LTD
A kind of CRISPR/Cas9 vector suitable for Clamella scutellaria fs482 and its construction method and application
ActiveCN112553238BHigh knockout efficiencyKnockout EfficientMicroorganism based processesEnzymesBiosynthetic genesSecondary metabolite
The invention discloses a CRISPR / Cas9 vector suitable for C. shieldia FS482 and its construction method and application. For the first time, the present invention utilizes the novel CRISPR / Cas9 system after promoter optimization to construct a recombinant Pseudomonas shieldiae FS482 strain in which the biosynthesis gene of the new diterpenoid skeleton compound has been knocked out, and establishes a high-efficiency C. The CRISPR / Cas9 gene knockout system of FS482 lays a molecular biological foundation for the elucidation of the biosynthetic mechanism of new active secondary metabolites of Pseudomonas FS482 and the acquisition of more new secondary metabolites with significant biological activity.
Owner:GUANGDONG INST OF MICROBIOLOGY GUANGDONG DETECTION CENT OF MICROBIOLOGY
A method for knocking out the chromosome of Saccharomyces cerevisiae
ActiveCN107858346BKnockout simpleKnockout EfficientHydrolasesStable introduction of DNABiotechnologyYeast chromosome
The invention relates to the technical field of biology, and specifically discloses a method for knocking down brewing yeast chromosome. The method is characterized in that the complete yeast chromosome is cut according to the CRISPR / Cas9 base technology; a specificity target close to centromere is selected; a corresponding guide RNA is designed to guide Cas9 protein to generate an incision closeto the centromere, thus realizing the knocking down of the whole chromosome. Compared with the conventional method for losing the whole chromosome in manners such as inducing through a Ga1 promoter, and reductional division, the method has the advantages that the brewing yeast chromosome is simply, efficiently and quickly cut; the cross exchange of sister chromatids can be avoided; the chromosome-knocked-down homozygous brewing yeast diploid strain can be obtained.
Owner:TIANJIN UNIV
CRISPR-Cas9-specific method for knocking out porcine FGL2 gene and sgRNA for specifically targeting FGL2 gene
ActiveCN105518139BKnockout fastKnockout precisionFermentationVector-based foreign material introductionBase JGenetics
The invention discloses a method for specifically knocking out the pig FGL2 gene by using CRISPR-Cas9 and an sgRNA for specifically targeting the FGL2 gene. The target sequence of the sgRNA specifically targeting the FGL2 gene of the present invention on the FGL2 gene conforms to the sequence arrangement rule of 5'-N(20)NGG-3', wherein N(20) represents 20 consecutive bases, wherein each Each N represents A or T or C or G; the target sequence on the FGL2 gene is located in the exon coding region of the FGL2 gene; the target sequence on the FGL2 gene is unique. The sgRNA of the present invention is used in the method for specifically knocking out the pig FGL2 gene by CRISPR-Cas9, which can quickly, accurately, efficiently and specifically knock out the pig FGL2 gene, and effectively solve the long period and high cost of constructing FGL2 gene knockout pigs The problem.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com