Kit for knocking out glioblastoma DHC2 gene
A glioblastoma and kit technology, applied in the field of alleles, can solve the problems of low editing efficiency, low off-target effect, low knockout efficiency, large workload, etc., and achieve high gene editing efficiency, strong specificity, and off-target rate. low effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
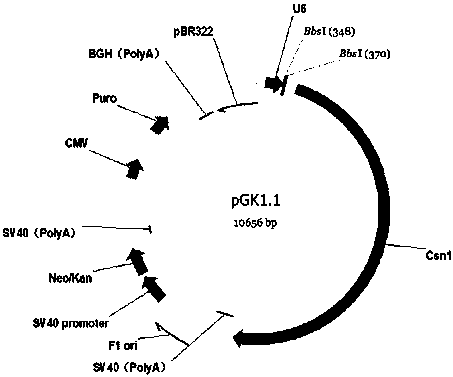
[0036] Example 1: Construction of the kit containing knockout plasmid according to the present invention
[0037] 1. Target site oligo DNA (oligo DNA) synthesis, and add adapters.
[0038] Using the online tool website (http: / / crispr.mit.edu / ), select the CDS region common to all transcripts of the DHC2 gene (gene ID: 79659), and find the third exon where the CDS region is located for the target site Design multiple gRNA sequences for knockout experiments.
[0039] Finally, 3 gRNA sequences were determined, the corresponding oligo DNA sequences were synthesized according to the 3 gRNA sequences, and additional bases were added to the head of the target sequence, CACC was added to the forward sequence, and AAAC (the first base of the target sequence) was added to the reverse sequence. The base must be G. If the first base of the selected target sequence is not G, you can add a G before the target sequence. The oligo DNA sequence corresponding to each gRNA is as follows:
[00...
Embodiment 2
[0084] Example 2: Using the kit of the present invention to stably knock out the DHC2 gene of the GBM cell line U87 cells
[0085] 1. To explore the conditions of electroporation of U87 cells
[0086] U87 cells were purchased from the ATCC cell bank in the United States. After the U87 cells were made into a uniform single cell suspension, a part of the cells was mixed and counted by trypan blue staining (the viable cell rate was >95% for electroporation), and the cell suspension was calculated. Concentration; take 3.0×10 6 The cells were suspended in a sterile tube and centrifuged at 1000 rpm for 4 min; the cell pellet was taken, a certain amount of DPBS was added to it, and the pEGFP-N1 plasmid (total 30ug, de-endotoxin treatment) was added after mixing, so that the final total system was at About 330ul; after mixing, add it into 3 electric shock cups with the electric rotary gun head respectively, so that the liquid level of the nozzle is raised, and no bubbles are formed i...
Embodiment 3
[0138] Example 3: Screening of knockout plasmids
[0139] This kit contains a unique knock-out plasmid, which has been verified by the inventor for many times. Compared with other knock-out plasmids, the knock-out plasmid contained in this kit has the characteristics of low off-target rate and strong specificity; The inventors have repeatedly verified that according to the method provided by the kit, the DHC2 two alleles can be stably knocked out, and the gene editing efficiency is extremely high.
[0140] The following table lists other gRNA sequences designed and tested by the inventor and the editing of the DHC2 gene, but the experimental results are not satisfactory.
[0141] Table 1. Other gRNA sequences and editing of DHC2 gene
[0142]
[0143] To sum up, the inventors unexpectedly found knockout plasmids with surprising effects and their combinations in experiments, and thus propose the kit for knocking out DHC2 gene in glioblastoma according to the present inventi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com