A kind of CRISPR/Cas9 vector suitable for Clamella scutellaria fs482 and its construction method and application
A technology of FS482, construction method, applied in the fields of biochemistry and molecular biology, which can solve problems such as low knockout efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Construction of a knockout vector targeting new backbone diterpenoid biosynthesis genes
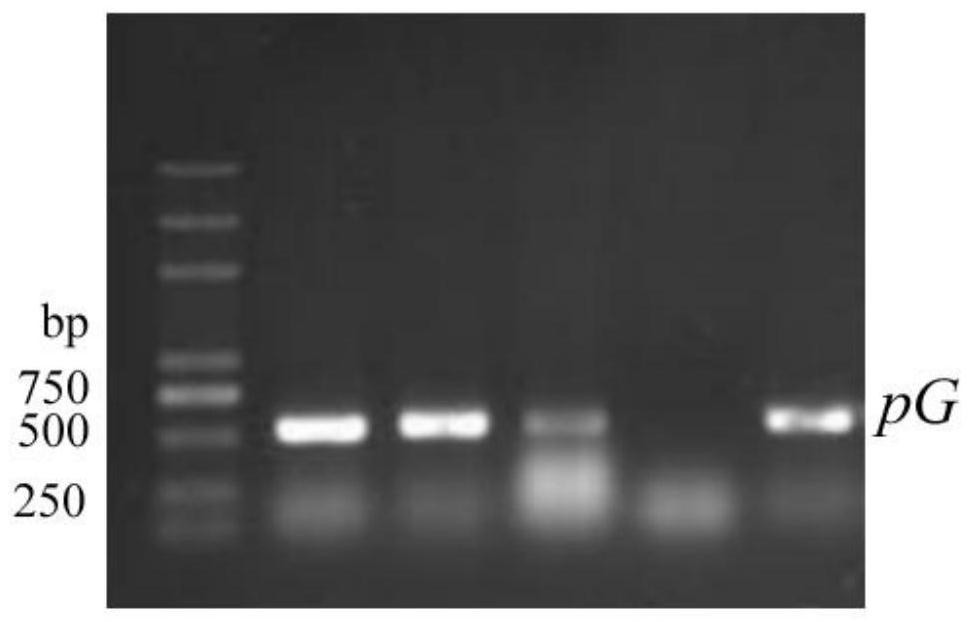
[0032] On the basis of the original pFC332 plasmid, pFC332 was PacI Single-enzyme digestion, homology arm sequences were designed on both sides of the Cas9 gene promoter of the pFC332 vector, combined with Escherichia coli ( Dichotomyces cejpii ) FS110 promoter pG base sequence (pG promoter was confirmed by in vitro luciferase transcription activity test and hygromycin resistance gene screening yeast experiment to have stronger transcription activity than pgpdA promoter), design primer sequence pG-F: CATCTAGAGGGCCCGCTTAATgaggaccgctcgtcccctat and pG-R: ctagtaatgcgtagaggtgcgtctgccatggtgatgggac; Dichotomyces cejpii) The FS110 genome was used as the template, and the above-mentioned pG-F and pG-R were used as primers to amplify the pG promoter sequence. PCR program: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 10 s, annealing at 55°C for 15 s, extension ...
Embodiment 2
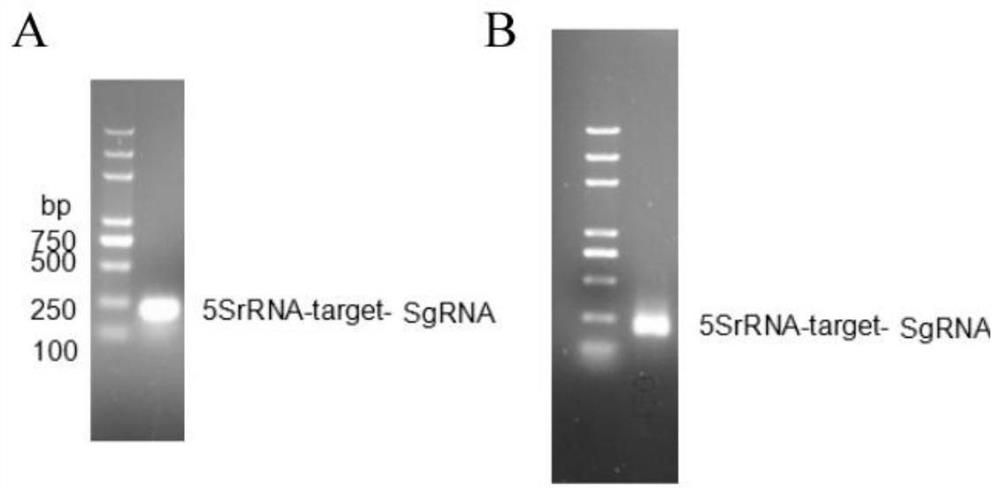
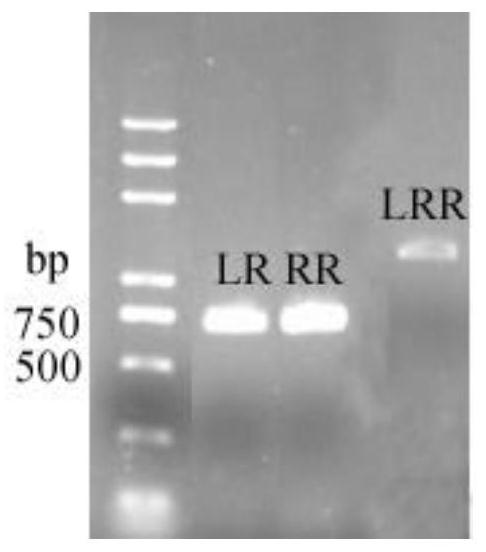
[0039] P. hawaiiense Knockout of P450 Gene Biosynthesis Gene in FS482:
[0040] foreign gene introduction P. hawaiiense The FS482 protoplast method is as follows:
[0041] (1) P. hawaiiense The preparation method of FS482 protoplasts is as follows:
[0042] Pick the right amount P. hawaiiense The hyphae of FS482 were inoculated into 200 mL PDA liquid medium and cultured at 30 °C and 180 r / min for 7 days. Filter the bacterial solution with two layers of gauze, select 2 g (wet weight) of well-grown bacterial balls in a 50 mL centrifuge tube, and wash twice with PBS buffer to fully wash away the residual PDA medium. Weigh 0.15 g of lysozyme (Sigma, USA) and dissolve it in 20 mL of KC buffer (0.6 M KCl, 0.05 M CaCl 2 ), and filtered through a 0.22 μm membrane filter, and added to the washed bacteria balls. Lysate at 28°C and 68 rpm for about 3 hours. Filter the lysate with a 200-mesh filter, filter the hyphae, filter again with 8 layers of sterile lens paper, centrifug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com