sgRNA sequence for specifically targeting arabidopsis ILK2 gene and application of sgRNA sequence
A DNA sequence, Arabidopsis technology, applied in the field of genetic engineering, can solve problems such as hindering wide application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Reality Example 1, Obtaining the Target Sequence of the Arabidopsis ILK2 Gene
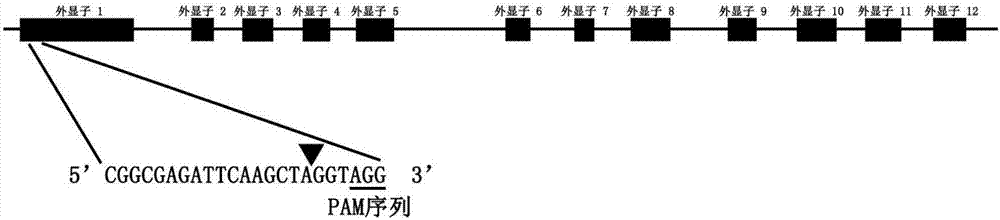
[0028] Obtain the coding region sequence of the first exon of the ILK2 gene on the Phytozome website (https: / / phytozome.jgi.doe.gov / pz / portal.html), and determine the target sequence according to the principle of sgRNA designed for knockout , located in the first exon, see figure 1 , the sequence is as follows:
[0029] SEQ ID NO.3: 5'-CGGCGAGATTCAAGCTAGGTAGG-3'
[0030] The sgRNA sequence targeting to edit this DNA sequence is as follows:
[0031] SEQ ID NO.1: 5'-CGGCGAGAUUCAAGCUAGGU-3'
[0032]The design principles of sgRNA are as follows:
[0033] 1. The length of sgRNA should generally be around 20nt
[0034] 2. The best content of GC% is 40%~60%,
[0035] 3. The binding position of the sgRNA targeting gene should be as close as possible to the downstream of ATG in the coding region of the gene, generally located in the first or second exon.
[0036] 4. The number of matches bet...
Embodiment 2
[0039] Embodiment 2, construction of Arabidopsis Cas9-ILK2KO vector
[0040] (1) sgRNA sequence: The 3-terminal PAM sequence AGG is removed from the target target sequence to obtain the sgRNA coding sequence SEQ ID NO. 2: 5'-CGGCGAGATTCAAGCTAGGT-3'.
[0041] (2) The reverse complementary sequence of the sgRNA coding sequence: the reverse complementary sequence of the sgRNA sequence was obtained to obtain the reverse complementary sequence SEQID NO.4: 5'-ACCTAGCTTGAATCTCGCCG-3'.
[0042] (3) Synthesis of single-stranded DNA oligonucleotides: according to the Bbs I restriction site of the selected Cas9 vector, 4 additional bases GATT are introduced at the 3 ends of the sgRNA sequence to obtain the sequence of SEQ ID NO.5; Four additional bases AAAC were introduced into the 5-end of the sgRNA reverse complementary sequence to obtain the sequence of SEQ ID NO.6. The sequences of SEQ ID NO.5 and SEQ ID NO.6 were handed over to Sangon Bioengineering (Shanghai) Co., Ltd. for artif...
Embodiment 3
[0068] Example 3, Cas9-ILK2KO transfected Agrobacterium EHA105
[0069] (1) Preparation of Agrobacterium Competent Cells:
[0070] Pick a single colony of Agrobacterium EHA105 and inoculate it in 5ml of YEB medium, cultivate overnight at 28°C on a shaker at 200rpm, inoculate it into 50ml of YEB medium at a ratio of 1:100 for expansion, and continue to cultivate at 28°C for about 6-7h until OD600=0.4 -0.6. Put the bacterial solution on ice for 30min; centrifuge at 5000rpm at 4°C for 5min, discard the supernatant, and suspend the bacteria in 10ml of 0.15M CaCl2; Suspended, distributed in 50 μl per tube, added sterile glycerol with a final concentration of 20%, and stored at -70°C.
[0071] (2) Transformation and identification of Agrobacterium:
[0072] Add 2 μl carrier Cas9-ILK2KO DNA to 50 μl Agrobacterium competent, mix well, ice bath for 30 minutes, freeze in liquid nitrogen for 4 minutes, 37°C water bath for 6 minutes, add 1ml YEB medium, 28°C, 200rpm shaking table for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com