Cell line for targeted knockout of pig GDPD2 gene based on CRISPR-Cas9 technology, and construction method of cell line
A cell line, IPEC-J2 technology, applied in the field of gene knockout model construction of porcine small intestinal epithelial cell line, can solve the problem that there is no small intestinal epithelial cell model
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
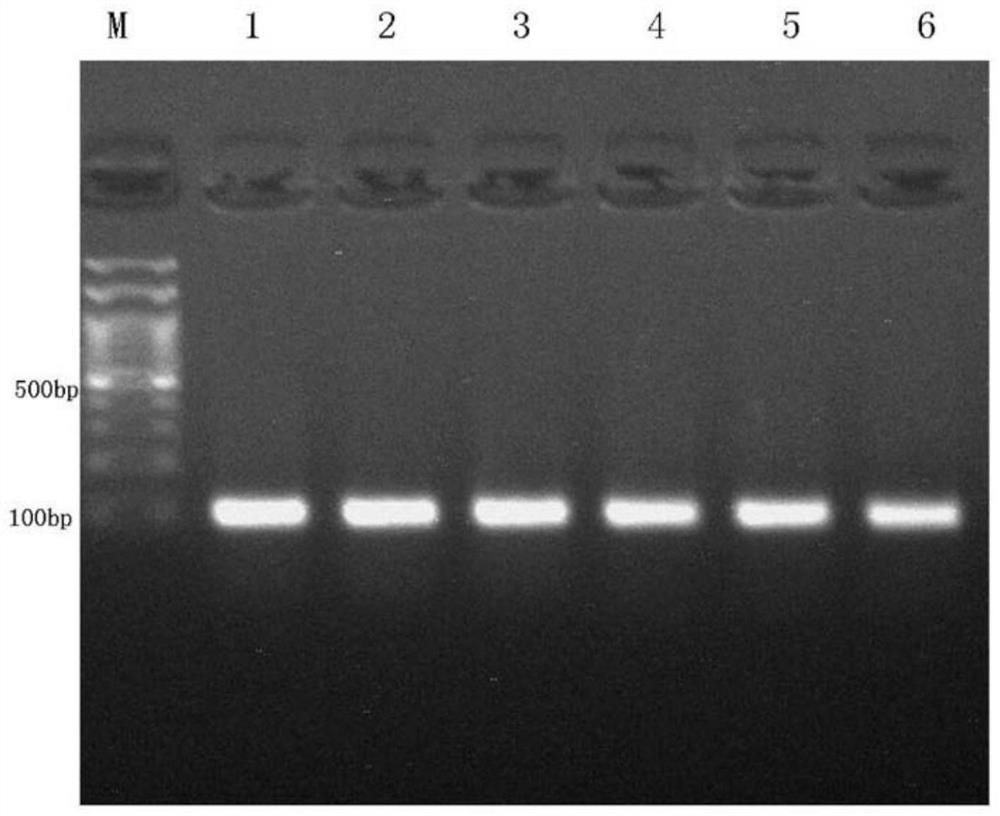
Examples
Embodiment 1
[0062] 1. Target site design and sgRNA sequence synthesis
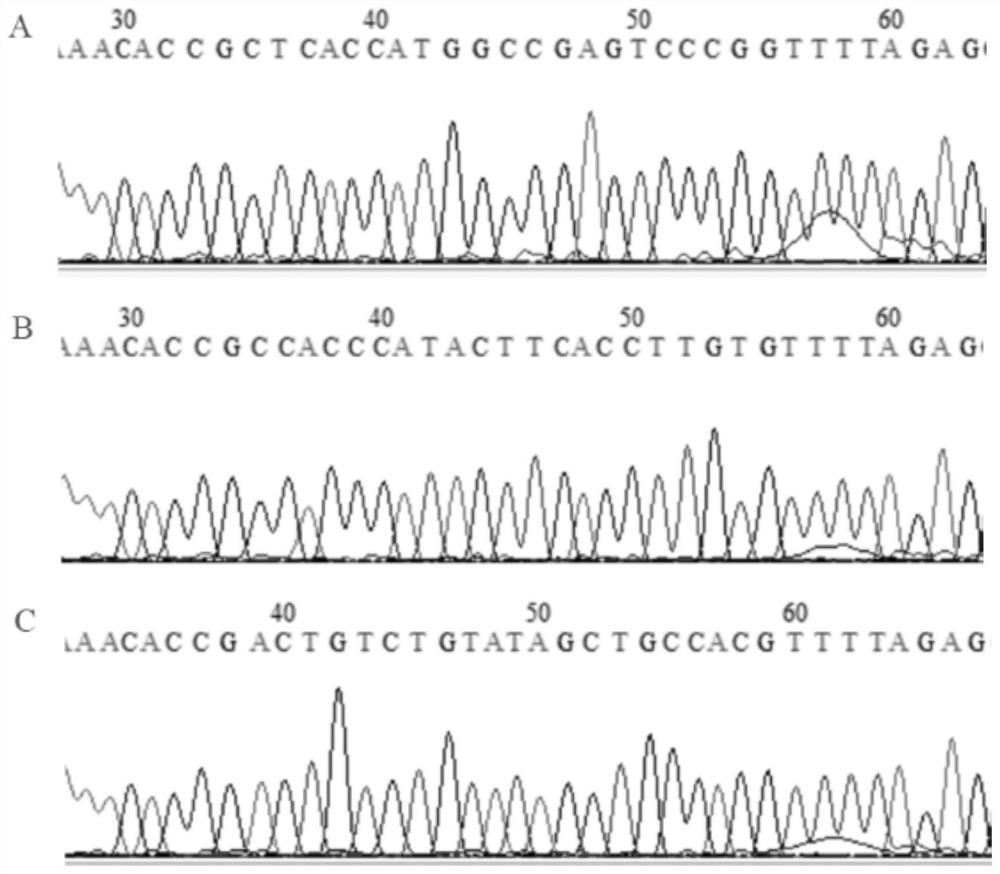
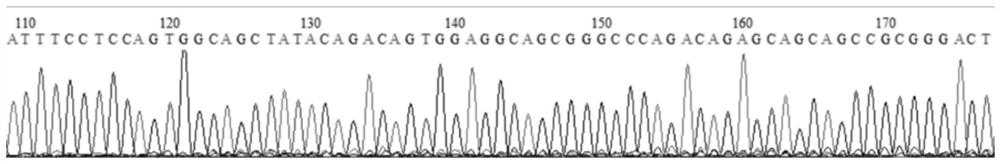
[0063] Find the CDS region of GDPD2 (Gene ID: 100516309) according to the NCBI database (https: / / www.ncbi.nlm.nih.gov / ), and design the knockout target site according to the first exon where the CDS region is located. Use CRISPRDesign (http: / / crispr.mit.edu / ) to design three sgRNA guide sequences:
[0064] sgRNA1: CTCACCATGGCCGAGTCCCGCGG
[0065] sgRNA2: CCAACAAGGTGAAGTATGGGTGG
[0066] sgRNA3: ACTGTCTGTATAGCTGCCACTGG
[0067] Add CACC at the 5' end of the sgRNA guide sequence and remove the PAM (NGG) sequence at the 3' end. If the first base at the 5' end of the sgRNA guide sequence is not G, add CACCG to form a positive-strand sgRNA sequence; the reverse of the sgRNA guide sequence The base AAAC is added to the 5' end of the complementary sequence to form a negative-strand sgRNA sequence. The three pairs of positive and negative strand sgRNA sequences are as follows:
[0068] GDPD2-1F: CACCGCTCACCATGGCCGAGTCCCG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com