Monocotyledon plant gene knockout vector based on CRISPR/Cas9 technology, and applications thereof
A monocotyledonous plant, gene knockout technology, applied in the biological field, can solve the problems of long cycle, inability to meet high-throughput plant genes, high cost, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
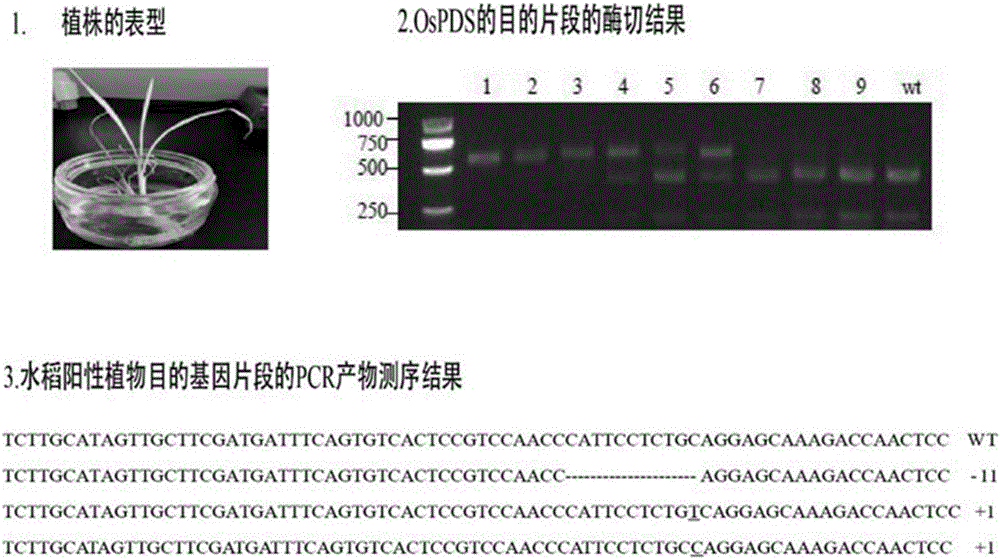
[0040] Example 1: Knockout and verification of rice albino gene
[0041] 1. Construction of rice albinism gene OsPDS knockout vector
[0042] 1. Design a special target sequence according to the rice albinism gene OsPDS sequence and sgRNA design tool, the sequence is as follows:
[0043] OsPDS-oligo1: GGCAGTTGGTCTTTGCTCCTGCAG (SEQ ID NO: 7)
[0044] OsPDS-oligo2: AAACCTGCAGGAGCAAAGACCAAC (SEQ ID NO: 8)
[0045] 2. Perform the degradation phosphorylation process of the designed target oligos according to the following conditions:
[0046] Prepare the reaction system in PCR tubes:
[0047]
[0048] Complete the reaction process on the PCR instrument according to the following procedures:
[0049]
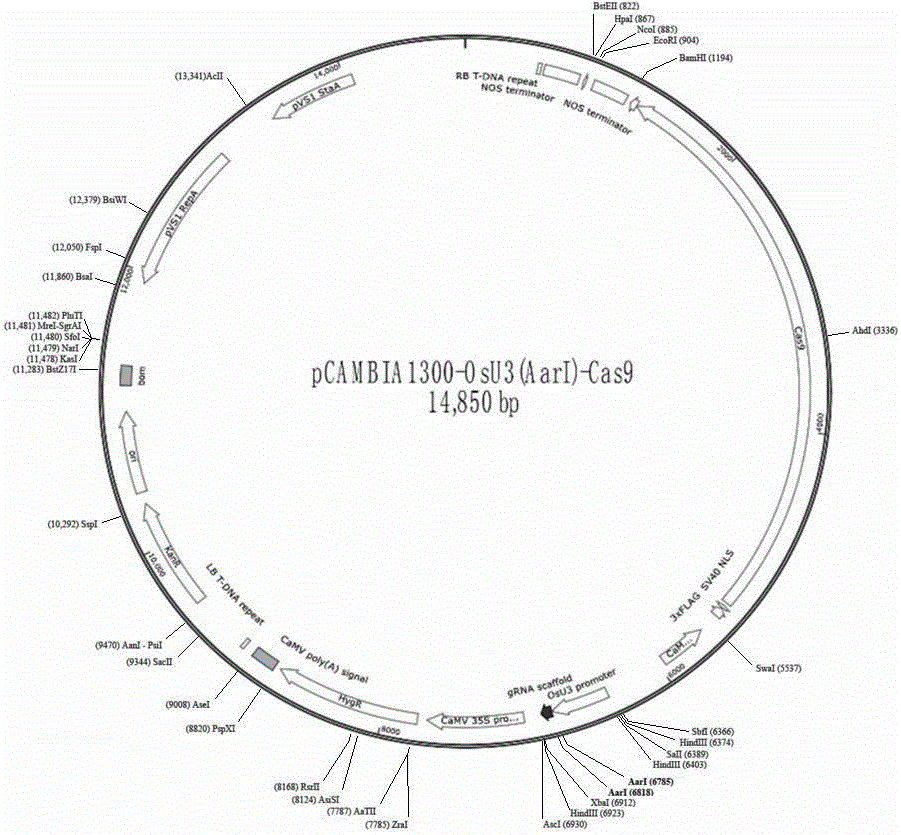
[0050] 3. Digest the pCAMBIA1300-OsU3(Aar I)-Cas9 plasmid with Aar I enzyme to obtain a linearized vector, and dephosphorylate it with CIP enzyme to avoid self-ligation.
[0051] 4. Dilute the annealed phosphorylated product 100 times, and take 2 μl for T4 ligation with the...
Embodiment 2
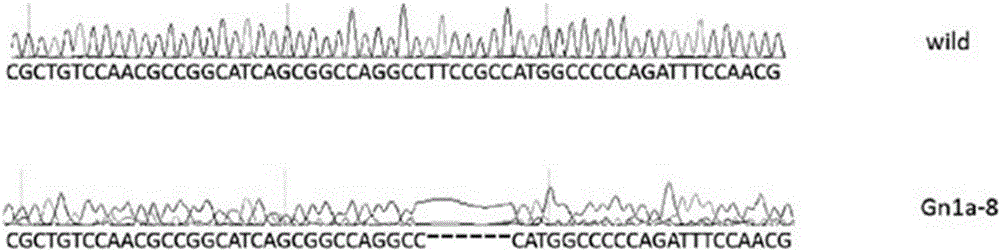
[0060] Example 2: Knockout and verification of rice Gn1a gene
[0061] 1. Design a special target sequence according to the rice gene Gn1a sequence and sgRNA design tool, the sequence is as follows:
[0062] Gn1a-2-oligo1: GGCAGCGGCCAGGCCTTCCGCCA (SEQ ID NO: 9)
[0063] Gn1a-2-oligo2:AAACTGGCGGAAGGCCTGGCCGC (SEQ ID NO: 10)
[0064] 2. According to the steps in Example 1, the knockout vector of Gn1a was successfully constructed.
[0065] 3. Transfer the knockout vector to Agrobacterium, and complete the whole tissue culture process to obtain transgenic positive seedlings for detection and verification.
[0066] 4. Detection of Gn1a transgenic positive plants: using Gn1a-F / R primers
[0067] Gn1a-F: 5'-GATTGATTGATTGATAATGAAGC-3' (SEQ ID NO: 11);
[0068] Gn1a-R: 5'-CCTATACCTTAATTACCTC-3' (SEQ ID NO: 12)
[0069] Perform PCR amplification of the target fragment, and sequence the PCR product. The PCR reaction conditions are 95°C, 30s; 55°C, 30s; 72°C, 40s; 35cycles.
[0070]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com