Construction method of vibrio parahaemolyticus gene efficient knockout plasmid
A technology of vibrio hemolyticus and plasmids, applied in the direction of microorganism-based methods, genetic engineering, plant genetic improvement, etc., to reduce the number and time of picking colonies, improve screening efficiency, and save time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1: Construction of knockout plasmid
[0048] Specific steps are as follows:
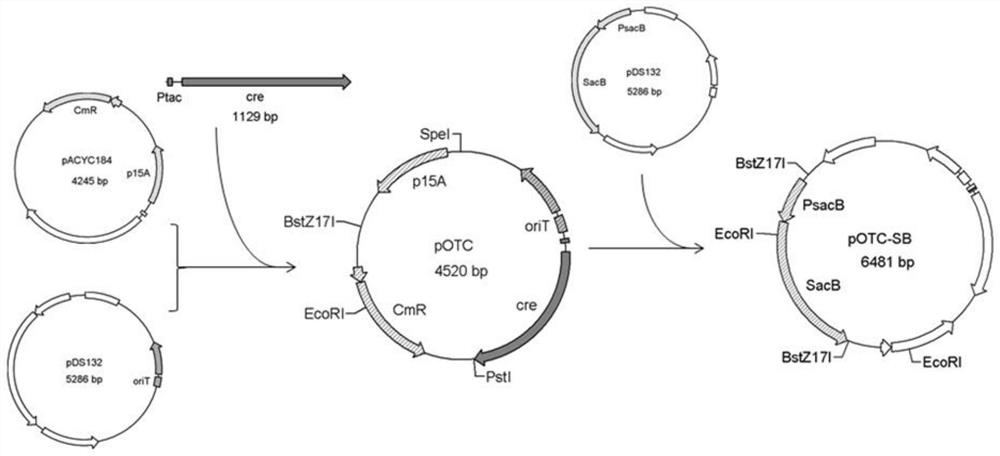
[0049] 1. Construction of plasmid pOTC
[0050] (1) Use primers Cm-p15A+ and Cm-p15A- to amplify the fragment containing the CmR and p15A genes from pACYC184. The PCR product was recovered using a PCR product recovery kit and digested with SpeI.
[0051] (2) Use primers traJ-oriT+ and traJ-oriT- to amplify traJ and oriT gene fragments from pDS132, use a PCR product recovery kit to purify the amplified DNA fragments, and digest them with SpeI.
[0052] (3) The cre gene fragment was amplified from pDTW109 using primers Ptac-cre+ and Ptac-cre-, and the PCR product was recovered using a PCR product recovery kit.
[0053] (4) Take 1 μg of the digested products in steps (1) and (2) respectively to configure a ligation system, carry out ligation at 4°C overnight, and treat at 70°C for 10 minutes to inactivate the ligase.
[0054] (5) Ligate the DNA fragment in step (3) and the ligation...
Embodiment 2
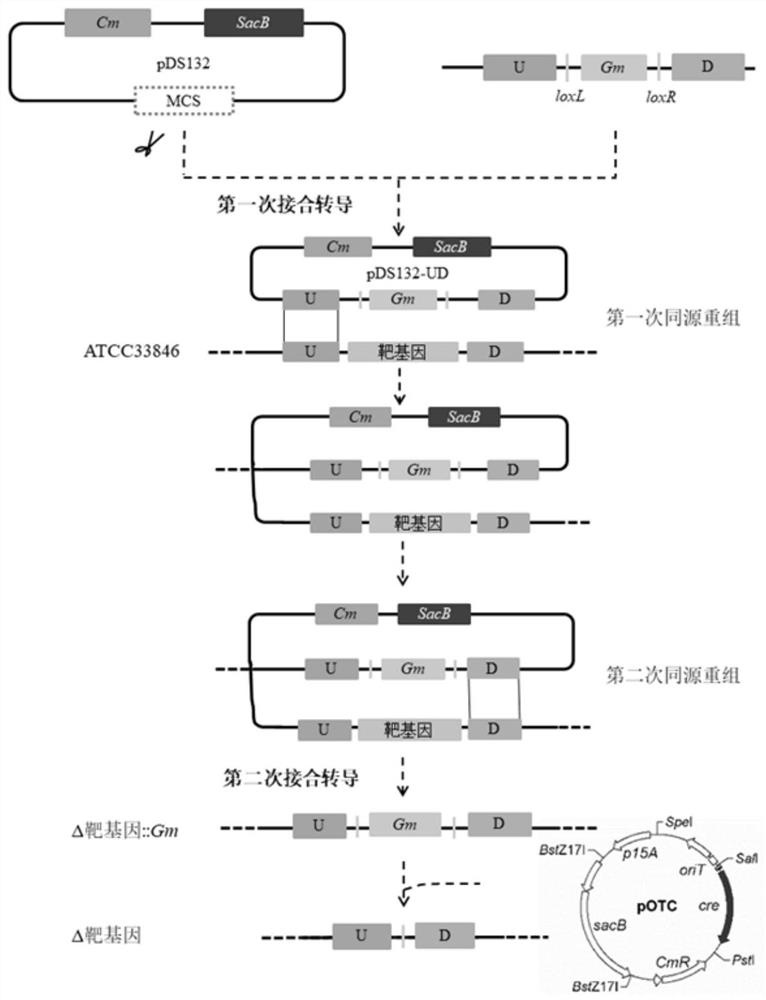
[0060] Example 2: Construction of Vibrio parahaemolyticus gene deletion mutant
[0061] Vibrio parahaemolyticus itself has a homologous recombination system. Taking advantage of this, the suicide plasmid containing the homology arm will be integrated into the genome after entering the cell. In this application, the accession numbers on NCBI of its sequence are VP_RS11975, VP_RS22195, VP_RS23020, VP_RS16800, VP_RS16465, VP_RS20840, VP_RS03765, VP_RS11205 genes, And rpoE whose accession number is VP_RS12550 on NCBI as an example, proves that the knockout method of the present invention can be effectively and successfully implemented.
[0062] Specific steps are as follows:
[0063] 1. Construction of gene knockout strains
[0064] (1) Use the upstream and downstream homology arm primers of VP_RS11975, VP_RS22195, VP_RS23020, VP_RS16800, VP_RS16465, VP_RS20840, VP_RS03765, VP_RS11205, and rpoE in Table 1 to perform PCR amplification to obtain the upstream and downstream homolog...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com