Construction method of primate miRNA-122 knockout model, primate liver cancer model and application thereof
A technology of miRNA-122 and primates, applied in botany equipment and methods, biochemical equipment and methods, applications, etc., can solve the problems of unsuitable liver cancer drug screening, unsuitable for human liver cancer mechanism research, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
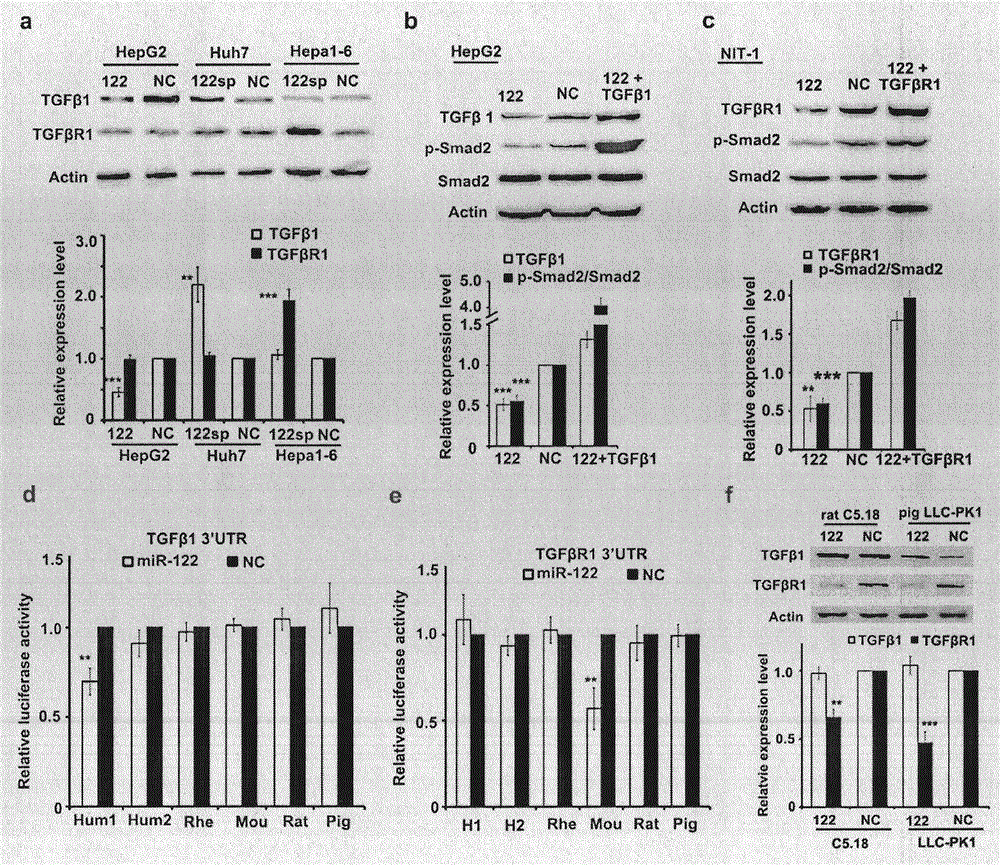
[0068] Refer to attached figure 1 . miR-122 directly targets TGFβ1 in human cells and TGFβR1 in mouse cells.
[0069] (a) Western blot analysis of TGFβ1 and TGFβR1 transfected with miR-122 overexpression plasmid (122), miR-122 sponge (122sp) and NC in HepG2, Huh7 and Hepa1-6 cells.
[0070] (b) Western blotting of HepG2 cells transfected with miR-122, NC and miR-122 and TGFβ1, which are respectively intracellular TGFβ1, Smad2 and p-Smad2, and the quantitative analysis results are on the right side of the figure, n=3.
[0071] (c) Western blot of NIT-1 cells transfected with miR-122, NC, and miR-122 and TGFβR1, which are intracellular TGFβR1, Smad2 and p-Smad2, respectively. Quantitative analysis results are on the right side of the figure, n=3.
[0072] (d) Luciferase activity was measured after transfecting the corresponding vector. The corresponding TGFβ1 3'UTR luciferase vector was constructed using the pGL plasmid. Hum: Human; Hum1: long 3'UTR; Hum2: short 3'UTR; Rhe...
Embodiment 2
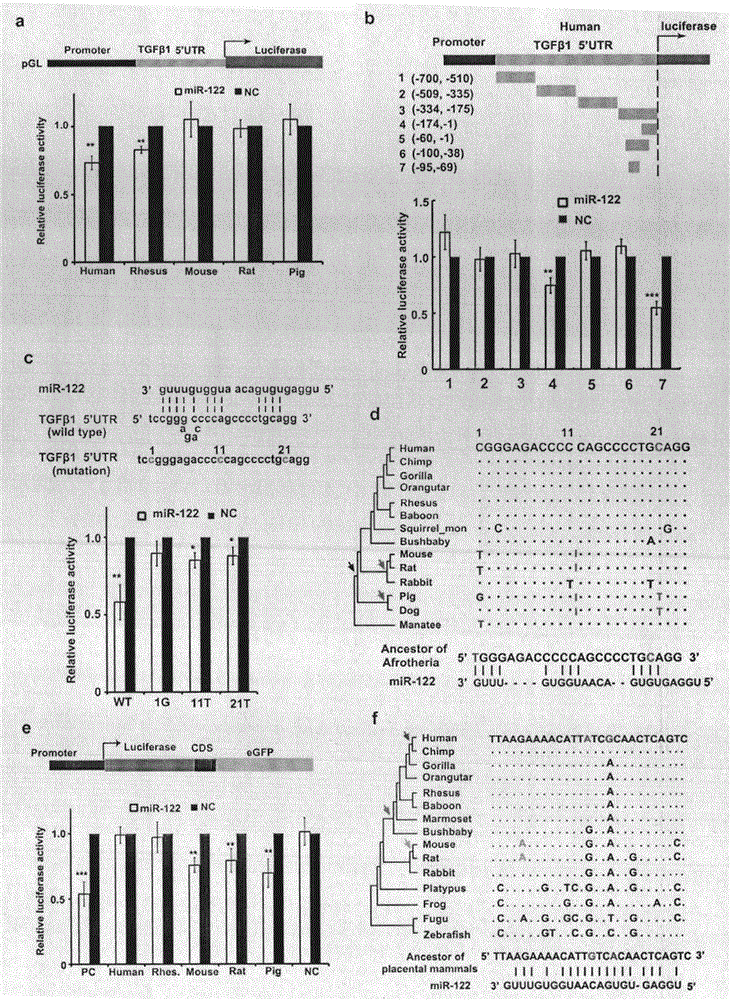
[0077] Refer to attached figure 2 . miR-122 targets TGFβ15'UTR in a non-canonical "seed region" base-pairing manner.
[0078] (a) Luciferase activity was measured after transfection of corresponding vectors. TGFβ15'UTR was ligated to the promoter region of pGL plasmid. n=6.
[0079] (b) Determination of luciferase activity after transfection of corresponding vectors. The human TGFβ1 5'UTR was truncated into 7 different segments and each segment was ligated into the promoter region of the pGL plasmid. n=6.
[0080] (c) Schematic representation of miR-122 acting on TGFβ15'UTR and mutant regions. The luciferase activities of different vectors are as follows.
[0081] (d) Evolutionary trajectory of miR-122 at TGFβ15'UTR target positions in different animals. Acquisition of the miR-122 target occurred in the common ancestor of manatees with humans and other primates (black arrows), in pigs, dogs, rats, and mice due to several insertions between positions 11 and 12 bases r...
Embodiment 3
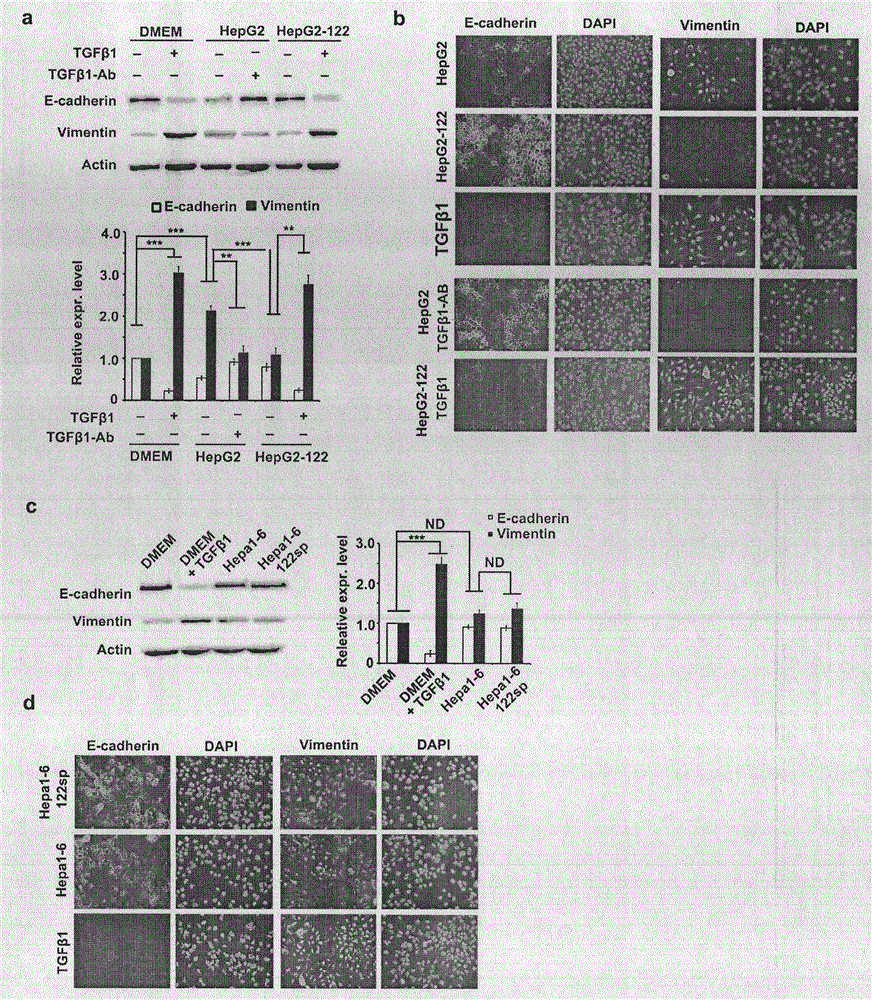
[0086] Refer to attached image 3 . Because targeting TGFβ1 or TGFβR1, respectively, resulted in differential effects of miR-122 on mesenchymal transition (EMT) in human and mouse cells.
[0087] (a) Western blot analysis of cadherin and vimentin in Huh7 cells after corresponding treatments. HepG2 and HepG2-122 represent the culture supernatant of HepG2 and HepG2-122 cells, respectively. Quantitative analysis results are on the right side of the figure, n=3.
[0088] (b) Nuclei were stained with DAPI using cadherin and vimentin as EMT labels in Huh7 cells. Scale bar: 50 μm.
[0089] (c) Western blot analysis of cadherin and vimentin in Huh7 cells after corresponding treatments. Hepa1-6 and Hepa1-6-122sp represent the culture supernatant of Hepa1-6 and Hepa1-6-122sp cells, respectively. Quantitative analysis results are on the right side of the figure, n=3.
[0090] (d) Nuclei were stained with DAPI using cadherin and vimentin as EMT labels for Huh7 cells. Scale bar: 50...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com