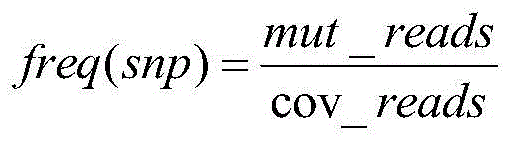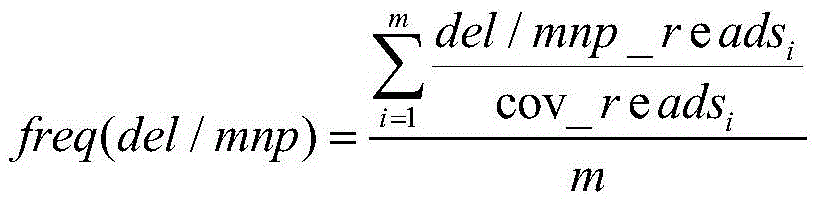High-throughput sequencing mutation detection result verifying method
A technology for detecting results and sequencing results, applied in the field of verification of high-throughput sequencing mutation detection results, which can solve problems such as false positives and missed detections
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0115] 1. Determining the Sensitivity of High-Throughput Sequencing Mutation Detection
[0116] Design the wild-type plasmid and the mutant plasmid. The mutant plasmid contains six commonly used mutation sites. The two plasmids are mixed in proportion to form a plasmid standard product with a mutation frequency of 0.5%. The library was built and sequenced, and each sample was repeated three times, and the raw data of the sequencing were counted. The results are shown in Table 1, which shows the mutation frequency of each sample at the above mutation site.
[0117] Table 1 Standard plasmid and wild-type plasmid sequencing results
[0118]
[0119] The t test was used to analyze the difference between the frequency of each mutation site in the mutant type and the wild type, and the results showed that the p value was 1.849×10 -15 , reaching a significant difference, which proves that high-throughput sequencing can accurately measure the mutation frequency at the level of 0.5...
Embodiment 2
[0121] 1. Obtain the mutation search results of high-throughput sequencing
[0122] The cfDNA extracted from the lung cancer patient No. 1253 was subjected to IonTorrent sequencing, and VariantCallerv3.0 supporting the IonTorrent sequencing platform was used to search for mutation information. The mutations in the found EGFR gene are shown in Table 2.
[0123] Table 2 EGFR gene mutation information of cfDNA of lung cancer patient number 1253
[0124] Chrome
position
GeneSym
type
mutInfo
cds_mut_syntax
aa_mut_syntax
COSM
VarFreq
Coverage
chr7
55242455
EGFR
SNPs
T>C
19
c.2225T>C
p.V742A
13183
4.06%
1108
chr7
55242470
EGFR
SNPs
T>C
19
c.2240T>C
p.L747S
26704
2.69%
930
chr7
55249054
EGFR
SNPs
C>T
20
c.2351C>T
p.S784F
13189
2.08%
240
chr7
55259515
EGFR
SNPs
T>G
21 ...
Embodiment 3
[0137] 1. Obtain the mutation search results of high-throughput sequencing
[0138] IonTorrent sequencing was carried out on the cfDNA extracted from lung cancer patients, and the mutation information was searched with VariantCallerv3.0 supporting the IonTorrent sequencing platform. ) mutations at 9 sites. VariantCallerv3.0 software reported mutation frequency as 0 for other sites.
[0139] Table 4 Mutation information found by VariantCallerv3.0
[0140]
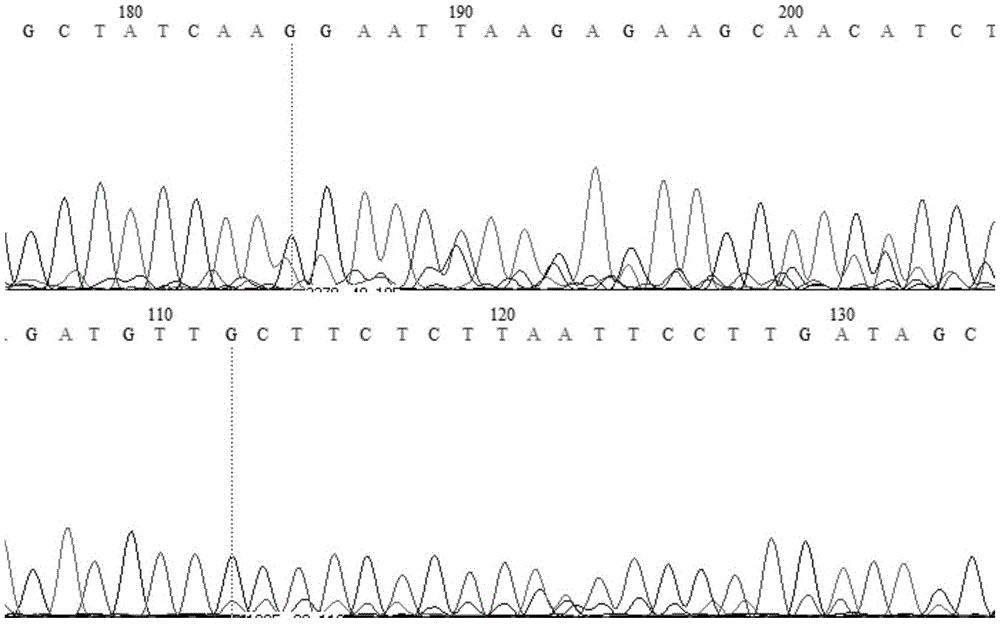
[0141] 2. Verify the hotspot mutation
[0142] Use the following formulas a and b to verify hotspot mutations not given in the mutation search results:
[0143] Formula a: f r e q ( s n p ) = m u t _ r e a d s cov ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com