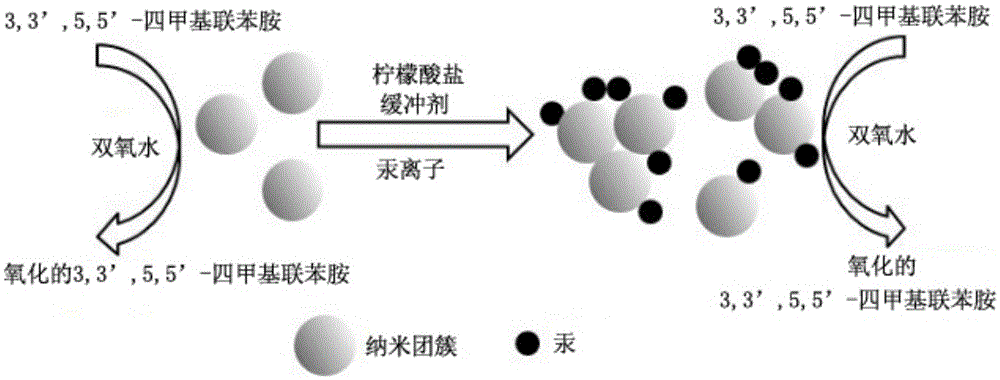
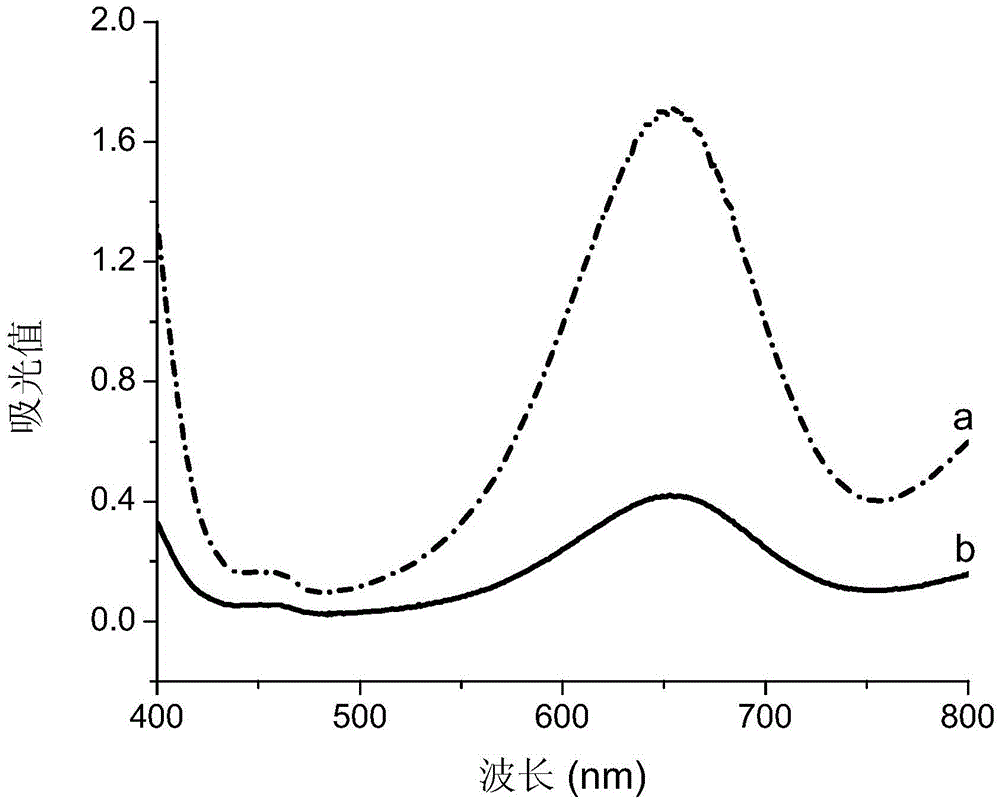
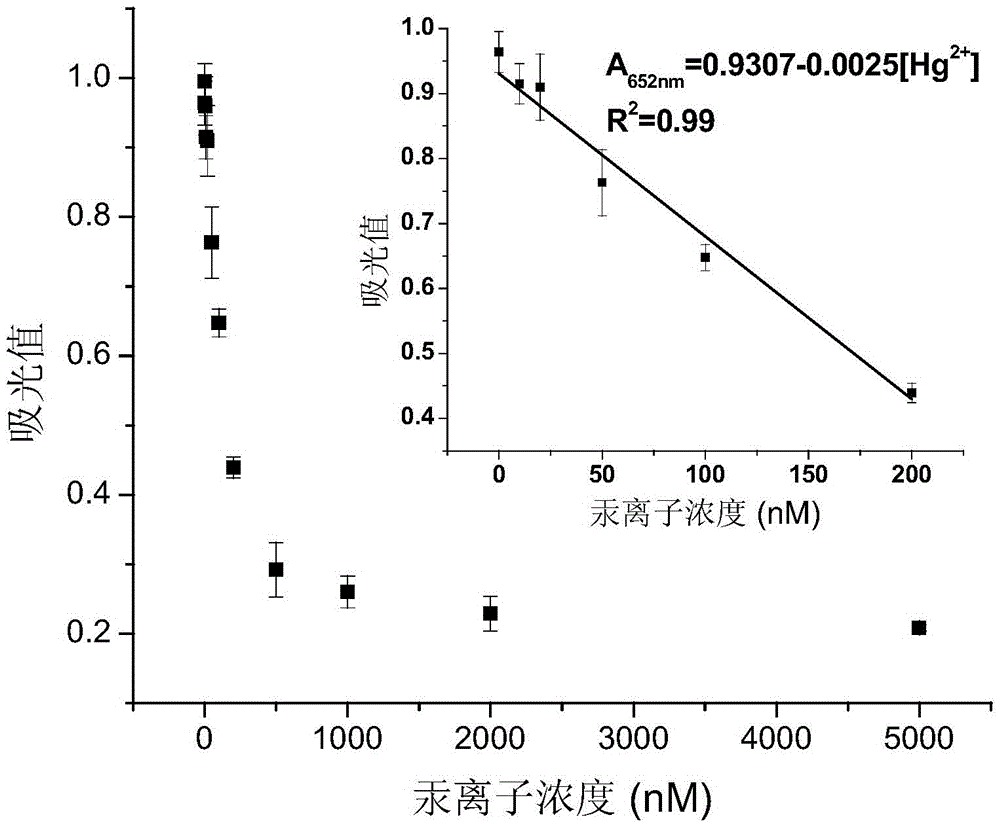
Mercury ion detection method
A detection method, mercury ion technology, applied in the field of nanotechnology, can solve the problems of strict use conditions, high cost, high cost, etc., and achieve the effect of high stability and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0050] Specifically, the preparation method of the DNA-Ag / Pt nanocluster solution includes:
[0051] Add 30 μL, 2.0 μM single-stranded nucleic acid (ssDNA), 5 μL, 150 μM silver nitrate solution and 12 μL, 125 μM potassium tetrachloroplatinate solution into a 200 μL PCR tube in sequence, and add 4 μL after reaction at 4°C for 30 min in the dark. A 5.0 mM sodium borohydride solution was shaken at 37 °C for 3 h to obtain a 2.0 μM DNA-Ag / Pt nanocluster solution.
Embodiment 1
[0054] (1) Synthesize DNA-Ag / PtNCs according to the reported references, and the specific steps are as follows:
[0055] Add 30 μL, 2.0 μM single-stranded nucleic acid, 5 μL, 150 μM silver nitrate solution and 12 μL, 125 μM potassium tetrachloroplatinate solution to a 200 μL PCR tube in sequence, and then add 4 μL, 5.0 mM potassium tetrachloroplatinate solution at 4°C for 30 min in the dark. The sodium borohydride solution was shaken at 37 °C for 3 h to obtain a 2.0 μM DNA-Ag / Pt nanocluster solution.
[0056] (2) Take 8 μL of the final mixed solution obtained in the step of diluting 2 times with ultrapure water, and add 2 μL of Tween-20 with a concentration of 0.005%-0.05% and 30 μL of sodium citrate with a pH value of 3.0 to 4.5 respectively. Buffer solution, 20 μL hydrogen peroxide with a concentration of 0.5M to 2.0M, 40 μL TMB solution with a concentration of 1.0 mM to 4.0 mM, and 100 μL of a series of standard Hg at different concentrations 2+ The solution was uniformly ...
Embodiment 2
[0060] (1) Synthesize DNA-Ag / PtNCs according to the reported references, and the specific steps are as follows:
[0061] Add 30 μL, 2.0 μM single-stranded nucleic acid (refer to Example 1, but not limited to this), 5 μL, 150 μM silver nitrate solution and 12 μL, 125 μM potassium tetrachloroplatinate solution into a 200 μL PCR tube, 4 ℃ After reacting in the dark for 30 min, 4 μL of 5.0 mM sodium borohydride solution was added, and the solution was shaken at 37° C. for 3 h to obtain a 2.0 μM DNA-Ag / Pt nanocluster solution.
[0062] (2) Step (1) of diluting 4 times with ultrapure water, 8 μL of the finally obtained mixed solution was added, and 2 μL of Tween-20 with a concentration of 0.005%-0.05% and 30 μL of sodium citrate with a pH value of 3.0 to 4.5 were added in turn. Buffer solution, 20 μL hydrogen peroxide with a concentration of 0.5M to 2.0M, 40 μL TMB solution with a concentration of 1.0 mM to 4.0 mM, and 100 μL of a series of standard Hg at different concentrations 2...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
| Concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com