Recombinant Salmonella expressing red fluorescent protein and its construction method
A red fluorescent protein and Salmonella technology, applied in the field of genetic engineering, can solve cumbersome problems, reduce false positives and save research funds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 The construction method of Salmonella delphi (SD-pFPV-mCherry) expressing red fluorescent protein
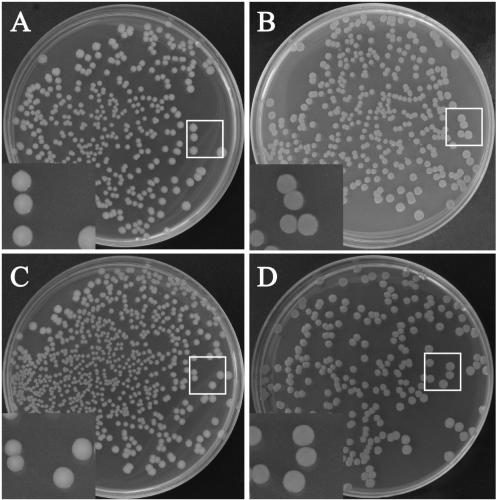
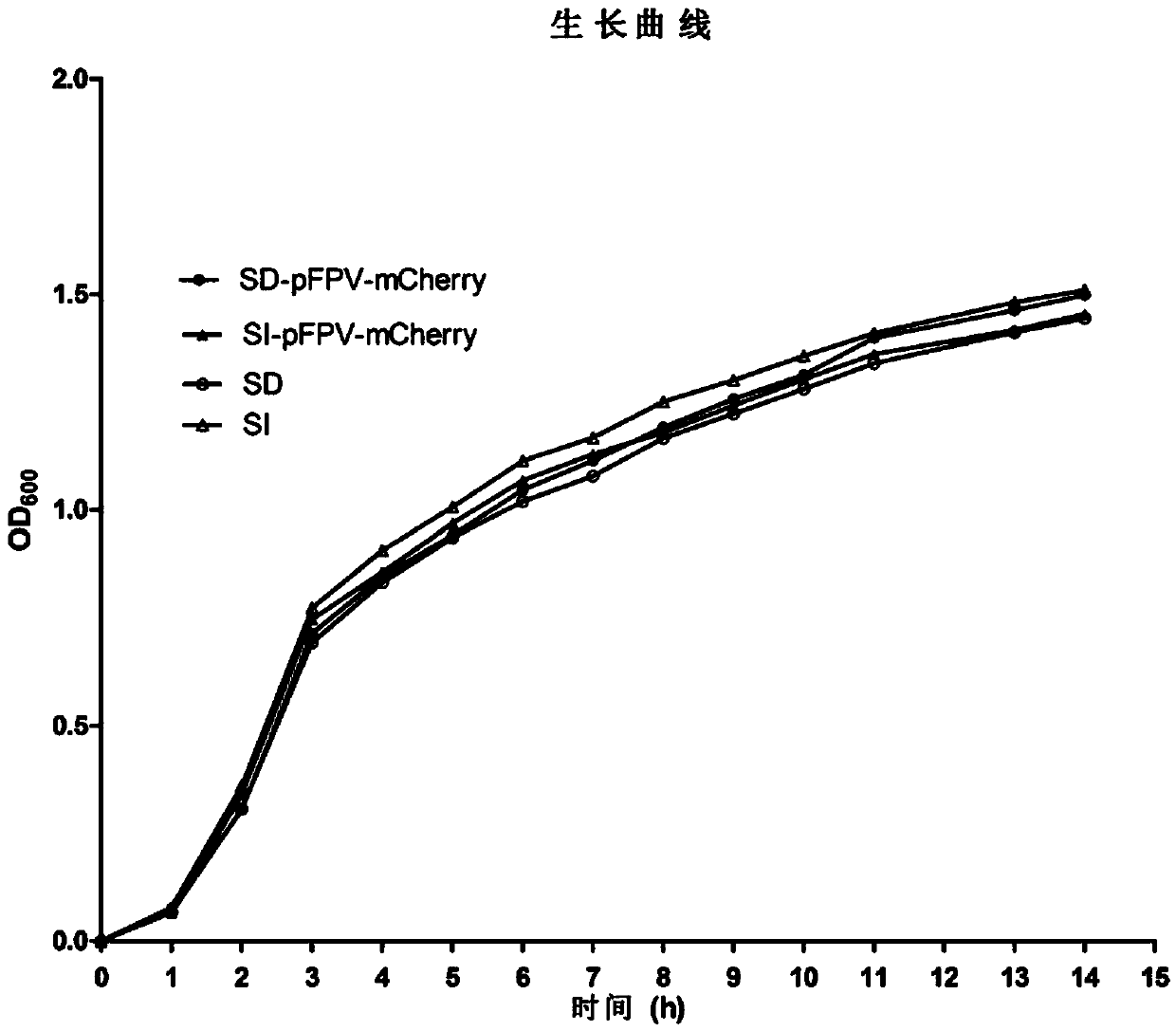
[0061] In this example, the clinically isolated Salmonella delphi is used as the transformation recipient, and the plasmid pFPV-mCherry inserted with the red fluorescent protein gene fragment is transformed into the bacteria by electric shock, thereby constructing Salmonella capable of expressing red fluorescence. Determine the genetic stability of the plasmid of the constructed recombinant strain, and detect whether there are significant differences in colony morphology, growth curve, and virulence factors compared with the parent strain, that is, to detect whether the transfer of the plasmid affects the normal biological characteristics of Salmonella. Technical route see figure 1 .
[0062] 1. Preparation of Competent Cells
[0063] (1) Pick a single colony of Salmonella from the XLD agar medium, inoculate it in a 50ml centrifuge tube containing 20ml LB cult...
Embodiment 2
[0113] Example 2 Construction method of Salmonella infantis (SI-pFPV-mCherry) expressing red fluorescent protein
[0114] The Salmonella infantis in this example is the same as the Salmonella delphi in Example 1, all of which are clinical isolates and belong to different serotypes. In this example, the preparation of Salmonella infantis competent cells, transformation by electric shock, identification of biological characteristics, etc. were carried out using the same steps as those in Example 1. Finally, Salmonella infantis capable of expressing red fluorescent protein was successfully constructed, and the plasmid was inherited after 100 generations of self-replication. The stability is more than 95%, which is similar to the recombinant Salmonella delbeis.
[0115] Salmonella expressing red fluorescent protein can be successfully constructed by adopting the method for constructing recombinant bacteria provided by the present invention. The recombinant bacteria have the same c...
Embodiment 3
[0116] Embodiment 3 Construction of recombinant Salmonella expressing green fluorescent protein
[0117] The plasmid used in this example was replaced by the plasmid pFPV-25.1 inserted with the green fluorescent protein gene fragment, and the construction method of the recombinant bacteria used the same steps as in Example 1. Constructed recombinant Salmonella delphi (SD-pFPV-25.1) and recombinant Salmonella infantis (SI-pFPV-25.1) expressing green fluorescent protein. There was no significant difference in the colony morphology, growth performance, and virulence factors of these two strains of recombinant bacteria, but the genetic stability of the plasmid was lower than 80% after 100 generations of self-replication ( Figure 7 ), it is not suitable for long-term experimental research, and can only be used for short-term experimental projects.
PUM
| Property | Measurement | Unit |
|---|---|---|
| strength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com