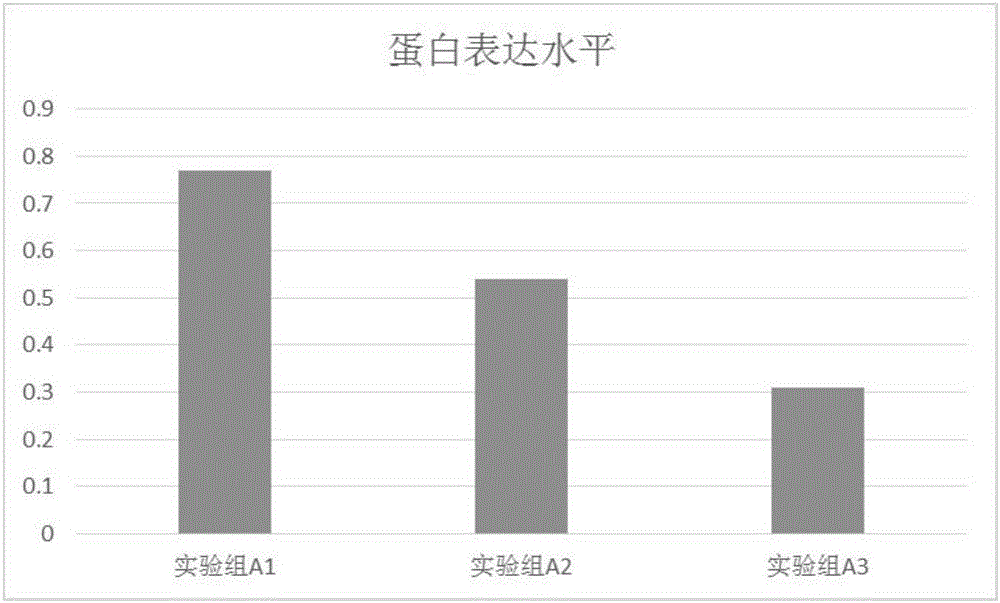
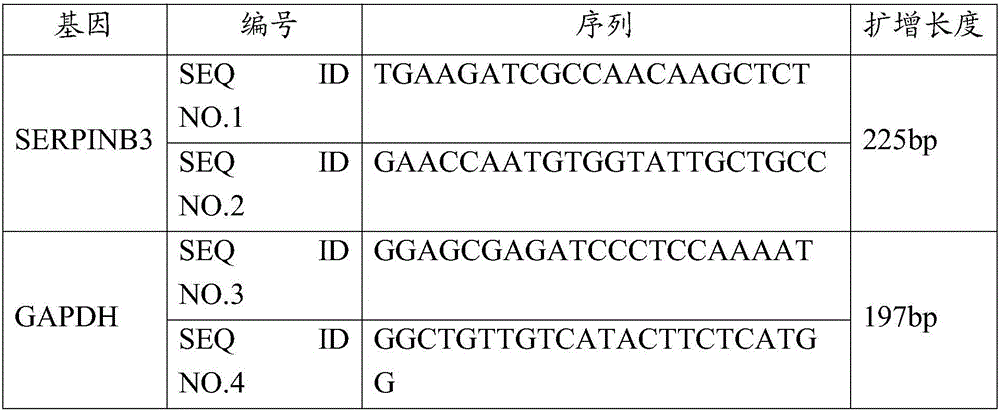
Application of SERPINB3 gene in preparing osteoarthritis diagnosis preparation
An osteoarthritis and gene technology, applied in the application field of human SERPINB3 gene in the preparation of osteoarthritis diagnostic preparations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Example 1 High-throughput sequencing screening of differentially expressed genes
[0070] 1. Sampling
[0071] From October 2012 to December 2015, patients with osteoarthritis who were treated in the Department of Orthopedics of Peking Union Medical College Hospital were collected. A total of 10 cases were collected as the case group. A total of 4 cases were collected from patients with other diseases who were hospitalized in the Department of Orthopedics during the same period as controls. The cartilage tissue samples of all subjects were obtained, numbered and stored in a -80°C low-temperature refrigerator.
[0072] The patients in the case group were all in line with the diagnostic criteria of knee OA and underwent knee arthroplasty; the control group were patients with meniscus injury and cruciate ligament injury who underwent arthroscopic surgery. According to the 1995 American Rheumatology Association diagnostic criteria for osteoarthritis, a patient with severe ...
Embodiment 2
[0094] Example 2 Verification of SERPINB3 gene expression in osteoarthritis patients and control cartilage tissues
[0095] 1. Materials
[0096] The cartilage tissues of 42 cases of osteoarthritis patients and 8 cases of control cartilage tissues were selected and grouped and numbered. The patients in the case group were all in line with the diagnostic criteria of knee OA and underwent knee arthroplasty; the control group were patients with meniscus injury and cruciate ligament injury who underwent arthroscopic surgery.
[0097] 2. Method
[0098] 2.1 Cartilage tissue is carried out total RNA is extracted, with the extraction method of embodiment 1.
[0099] 2.2 Synthesis of cDNA by reverse transcription
[0100] use IIIReverseTranscriptase (invitrogen, Cat. No. 18080-044) was used for cDNA reverse transcription, and the experimental operation was carried out according to the product manual. The specific operation was as follows:
[0101] Using a reverse transcription k...
Embodiment 3
[0119] Example 3 Papain-induced osteoarthritis model
[0120] There are many ways to establish animal models of OA, such as Hulth model, model of anterior cruciate ligament and meniscectomy, intra-articular injection of papain, ovariectomy, spontaneous formation of high-fat food feeding, joint immobilization, etc. Related studies have shown that injecting papain into the marrow joint or knee joint cavity of rabbits and guinea pigs can induce rapidly progressive osteoarthritis. Papain can decompose the proteoglycan in the cartilage matrix and promote its loss from the cartilage. The most significant changes in the cartilage of patients with osteoarthritis in the early stage are the increase of water and the decrease of proteoglycan. Therefore, the papain-induced OA model is consistent with human bone Arthritis is similar and is a better model for studying OA.
[0121] Experimental animals: 30 healthy male New Zealand white rabbits, 6 months old, weighing about 2kg. After the e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com