Image visualization method for detecting single molecule DNA duplication
A single-molecule, single-stranded technology, applied in the field of image visualization to detect single-molecule DNA replication, which can solve problems such as unsolvable description and characterization, lack of direct observation methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] The preparation of embodiment 1 single-stranded DNA-DNA origami
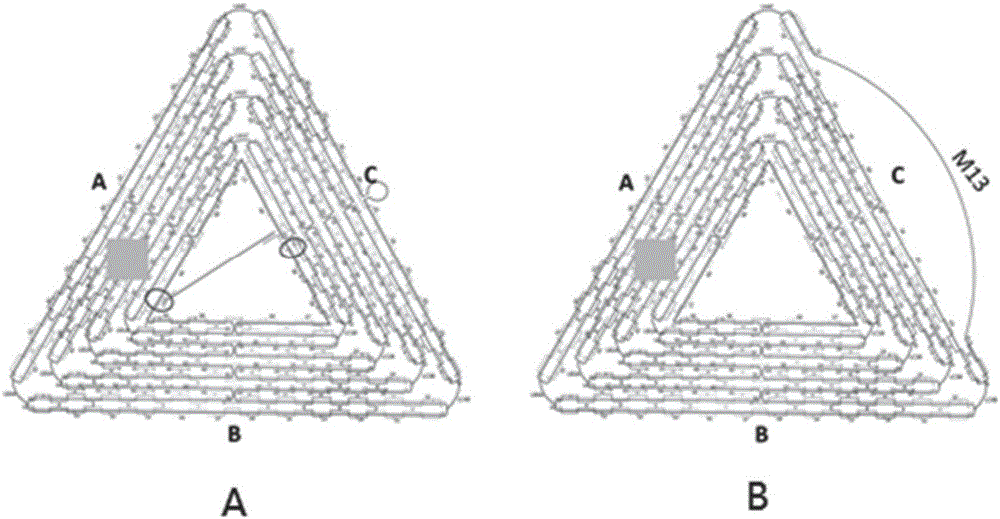
[0039] The hollow DNA origami designed in this example utilizes M13mp18DNA and staple single-strand self-assembly to form an equilateral triangle pattern. The preparation method of the equilateral triangle hollow DNA origami is as follows: the ring-shaped long scaffold single-stranded DNA M13mp18 and the two strands A17 and C33 are respectively The staple strand DNA collection ABCL of the sequence shown in SEQIDNO.1 and SEQIDNO.2 is replaced by the sequence table and mixed in TAE-Mg 2+ Buffer system (40mM Tris-acetic acid, 1mM EDTA, 12.5mM MgCl 2 , pH8.0), and then placed in a PCR instrument to anneal from 95°C to 20°C at an annealing rate of 0.1°C / 10s, wherein the two strands of M13mp18, A17 and C33 were replaced by the sequences listed in SEQ ID NO.1 and SEQ ID NO.2, respectively. The molar concentration ratio of the staple strand DNA collection ABCL of the sequence shown is 1:10, with TAE-Mg 2+ The b...
Embodiment 2
[0041] The preparation of embodiment 2 single-stranded DNA-DNA origami
[0042] The hollow DNA origami designed in this example uses M13mp18DNA and the staple chain DNA set ABCL to self-assemble to form an equilateral triangle pattern, wherein the staple chain part of the C side of the equilateral triangle is shortened, specifically, C08, C12, C16, C20, C23, C24, C26, C28, C30, C41, C45, C49, C53, C56, C59, C61, C63 and C64 were replaced by sequences such as sequence listing SEQIDNO.4, SEQIDNO.5, SEQIDNO. 6. SEQ ID NO.7, SEQ ID NO.8, SEQ ID NO.9, SEQ ID NO.10, SEQ ID NO.11, SEQ ID NO.12, SEQ ID NO.13, SEQ ID NO.14, SEQ ID NO.15, SEQ ID NO.16, SEQ ID NO.17, SEQ ID NO.18, The sequences shown in SEQIDNO.19, SEQIDNO.20 and SEQIDNO.21 make the corresponding part of the M13mp18 single-stranded DNA unpaired, forming a DNA strand on the outer edge to become a single-stranded DNA. The components and methods of other assembly systems are the same as in Example 1 , the structure diagram...
Embodiment 3
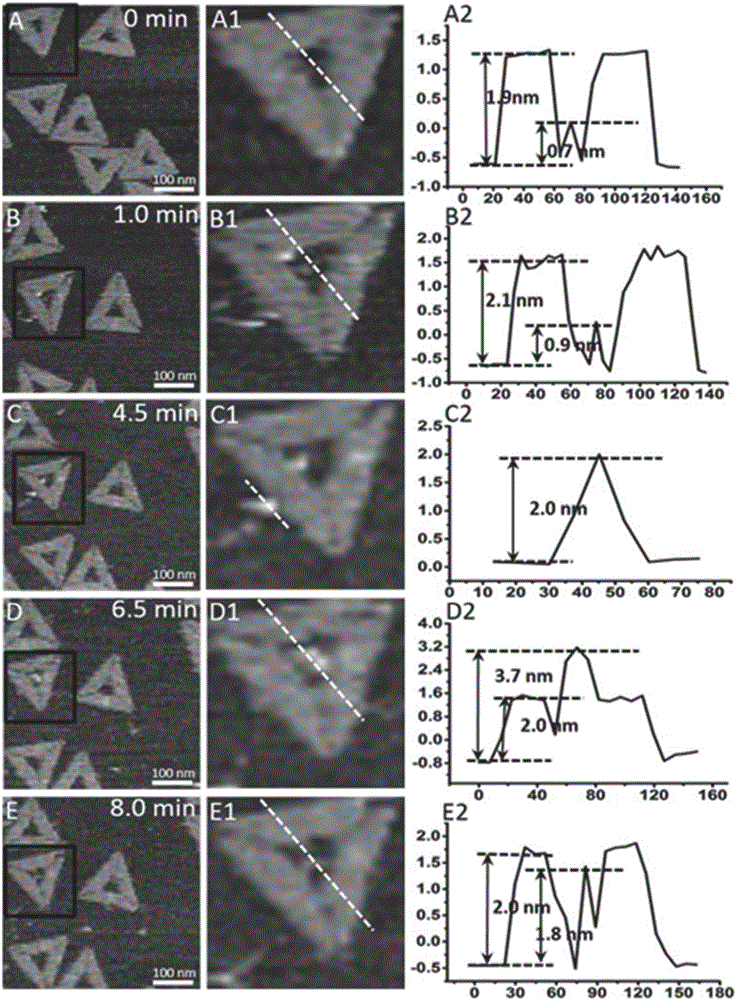
[0043] Example 3 Real-time dynamic image visualization detection and analysis of DNA replication by atomic force microscope
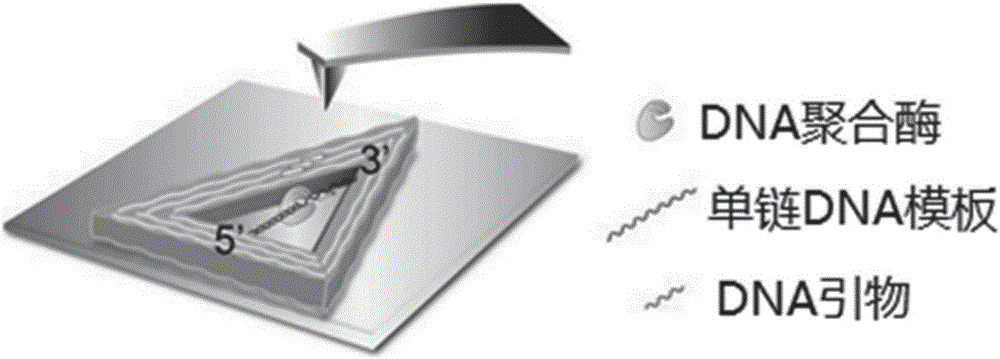
[0044] Add 3.0 μL of the triangular single-stranded DNA-hollow DNA origami prepared in Example 1 dropwise on the surface of the newly dissociated mica and let it stand for 3 minutes. After adding 30 μL of TAE buffer to the liquid pool, use an atomic force microscope to scan and image to obtain clear DNA Image of origami nanostructures and DNA template strands, modeled as figure 2 shown. Subsequently, a mixed solution containing 2 μL of DNA polymerase Klenow fragment at a concentration of 85 U / mL and 2 μL of dNTPs at a concentration of 62.5 μM was added to the liquid pool, and scanned and imaged immediately. Scanning adopts "J" scanning head, "tap" mode, and adjusts scanning parameters to form images in the small force area (F<200pN). Atomic force microscope images of samples were collected at 1 min, 4.5 min, 6.5 min and 8 min after adding the mixture...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Elastic coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com