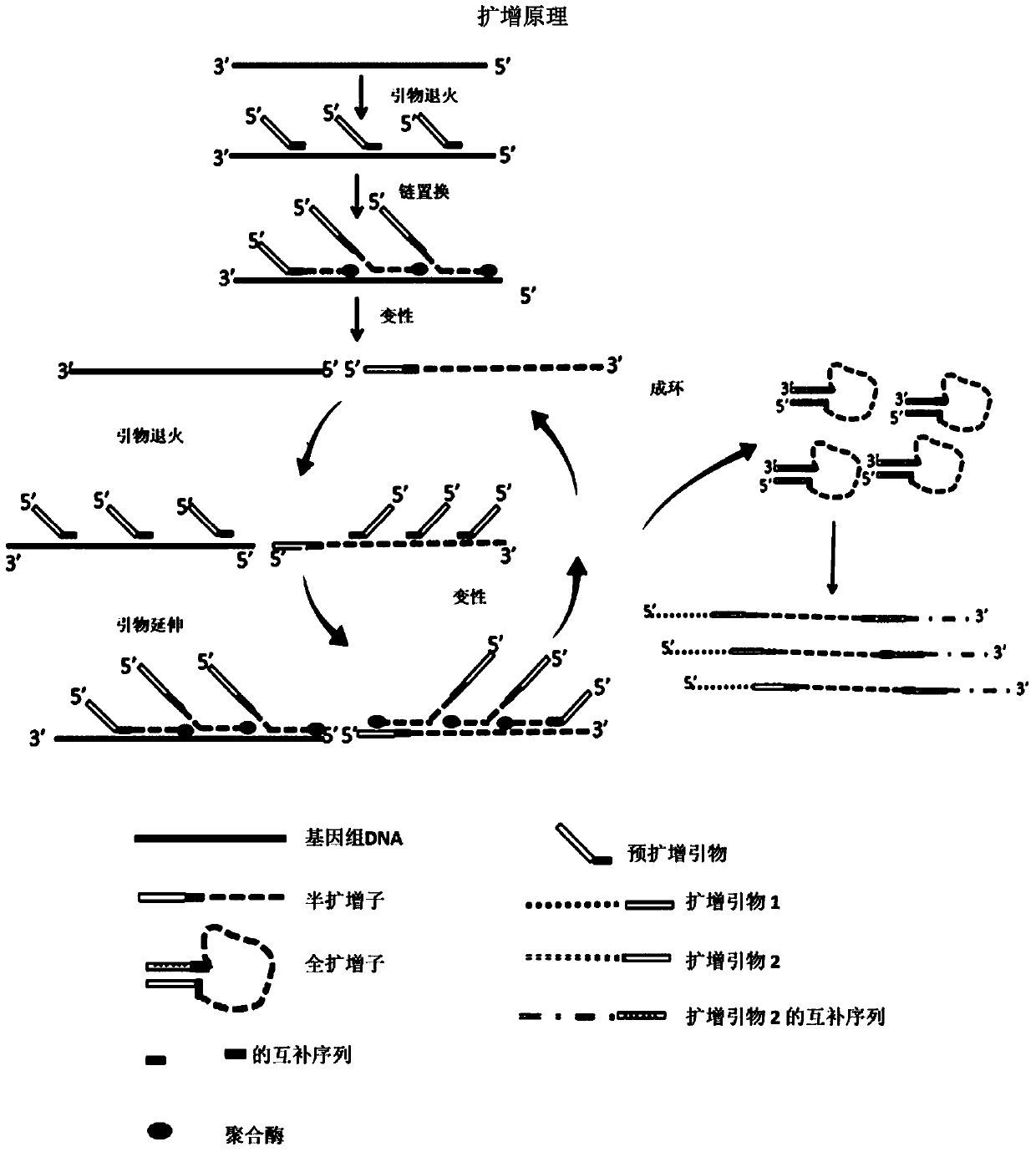
how to amplify dna
A technology for pre-amplification and amplification of products, applied in the field of DNA amplification, which can solve the problems of poor sequencing results, cumbersome operations, and long time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0186] Example 1: Preliminary verification of amplification effects using different linear amplification primer mixtures
[0187] a) Validation using standard genomic DNA samples
[0188] The standard genomic DNA is the genomic DNA of human cells extracted in advance. Dilute the standard genomic DNA with nuclease-free water to a DNA solution of 50 pg / μl, take 1 μl of the above solution (as a source of genomic DNA) and add it to a PCR tube, and add it to each experimental group as shown in Table 1. The primer mix and other relevant reagents, obtain the first reaction mixture (which contains Na + , Mg 2+ , Cl - , Tris-Cl, TritonX-100, dNTPs, Vent polymerase and primer mix).
[0189] linear amplification
[0190] For each primer combination / mixture, two experimental groups were used to conduct parallel experiments to ensure its accuracy, and the reaction mixture of each experimental group was placed in the following first temperature control program for reaction:
[019...
Embodiment 2
[0230] Example 2: Further verification of amplification effects using different linear amplification primer mixtures
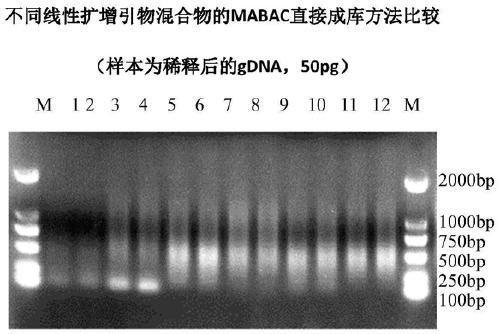
[0231] Human epidermal fibroblasts were isolated and lysed according to the method described in implementation 1b) to obtain single-cell genomic DNA, and the primer mix used in the experimental group 11 / 12 in Table 1 and the primer mix used in the experimental group 9 / 10 were used, respectively. The primer mixture was used for amplification, and 10 parallel experiments were performed for each primer mixture (represented as 1_1, 1_2...1_10 and 2_1, 2_2...2_10). Amplify according to the program described in embodiment 1a) and obtain the amplified product, and carry out gel electrophoresis detection to the amplified product, the electrophoresis detection result is as follows Image 6 shown. Wherein the concentration of the amplified product in the experimental group 2_1, 2_2...2_10 is slightly lower than the concentration of the amplified product in the experime...
Embodiment 3
[0239] Example 3: Detection of pathogenic sites and quality inspection primers
[0240] Pathogenic site detection
[0241] 35 disease-causing loci were randomly selected (see Table 8 below for the selected loci), and primers were designed. The selected pathogenic loci and their corresponding primers are shown in Table 8 and Table 9, respectively.
[0242] Table 8: 35 randomly selected disease-causing loci
[0243] Pathogenic site name chromosome location 1 SMN1-1 chr5 2 SMN1-2 chr5 3 SMN1-3 chr5 4 SMN1-4 chr5 5 SMN1-1R chr5 6 SMN1-2R chr5 7 SMN1-3R chr5 8 SMN1-4R chr5 9 PDS-IV15 chr7 10 PDS-EXON5 chr7 11 PDS-EXON7+8 chr7 12 PDS-EXON10 chr7 13 PDS-EXON17 chr7 14 PDS-EXON19 chr7 15 HBB3 chr11 16 HBB chr11 17 MMACHC chr1 18 HBA2 chr16 19 GJB2 chr13 20 GJB2-C796 chr13 21 ATP7B-8 chr13 22 PKHD1-3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com