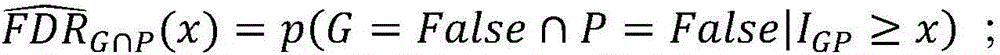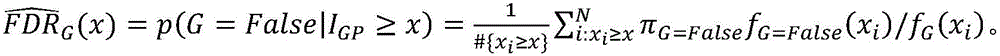Complete glycopeptide identifying method and system
A glycopeptide and complete technology, applied in the fields of glycoproteomics, mass spectrometry, and bioinformatics, can solve the problems of indistinguishability and poor reliability, and achieve the effects of accurate false discovery rate, improved reliability, and low computational complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
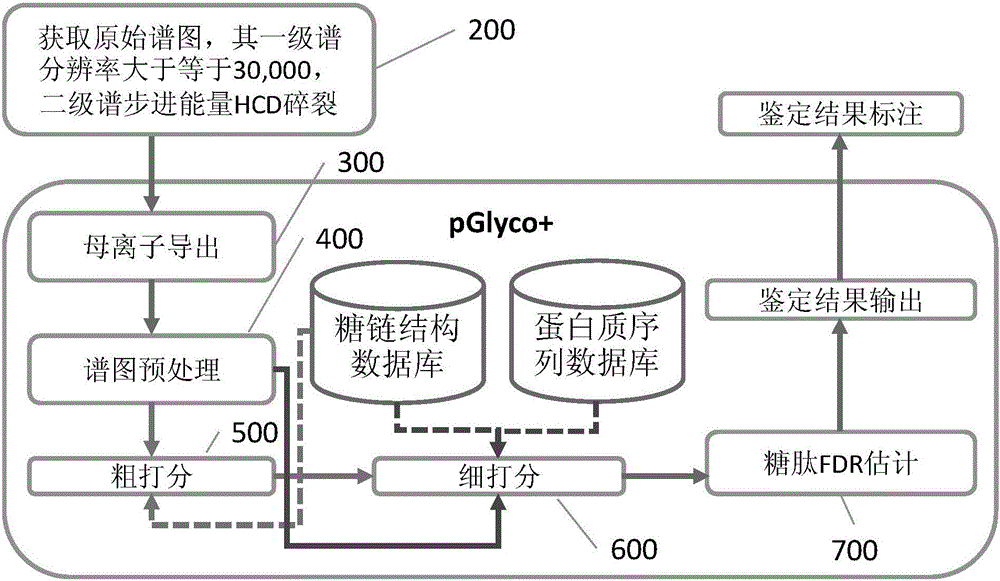
[0059] For ease of understanding, first give the meaning of some professional concepts involved in this article:
[0060] Glycosidic bond: a chemical bond formed by connecting monosaccharides and monosaccharides;
[0061] The reducing end of the sugar chain: refers to the end containing the free radical -CHO. In glycoproteomics, it refers to the end of the sugar chain that is connected to the peptide;
[0062] The reducing end ion of the sugar chain: after the glycosidic bond of the sugar chain is broken, the fragment ion at the reducing end is sometimes called the Y ion of the sugar chain
[0063] Y ion of glycopeptide: the reducing end ion of the sugar chain plus the peptide sequence of the complete glycopeptide;
[0064] B / y ion of glycopeptide: b / y ion of peptide part of glycopeptide, b / y ion does not contain any sugar chain structure;
[0065] Pentasaccharide core: fixed five monosaccharide structures generally present at the reducing end of N sugar chain, namely 2×HexNAc+3×Hex.
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com