SNP (simple nucleotide polymorphism) modular marker for rice low cadmium accumulation gene OsHMA3 and application thereof
A technology of molecular markers and low accumulation, applied in the field of molecular biology, can solve the problems of difficult and accurate determination of phenotypic values, few SNP markers, few main effect genes, etc., to achieve accurate prediction of cadmium content in rice grains, easy operation, and application value strong effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Development of SNP Molecular Marker of Rice Low Cadmium Accumulation Gene OsHMA3
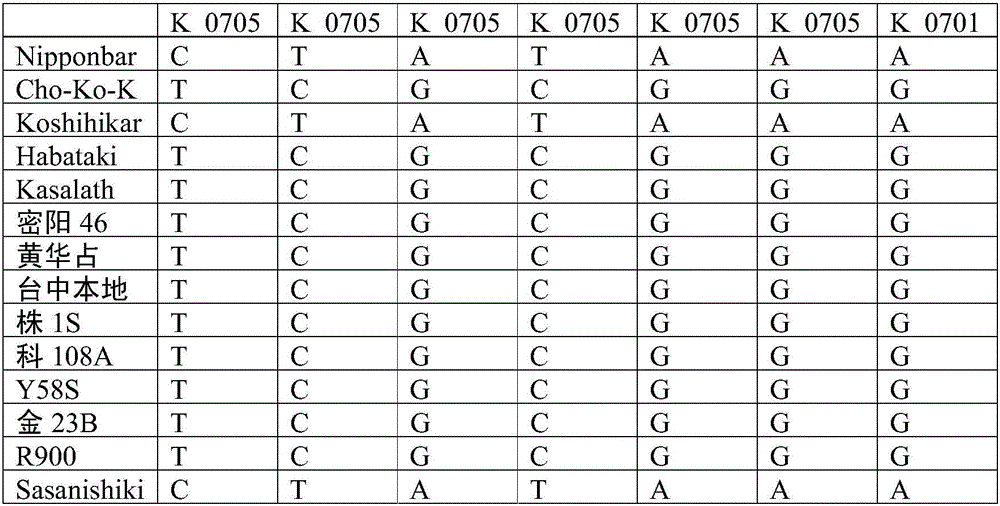
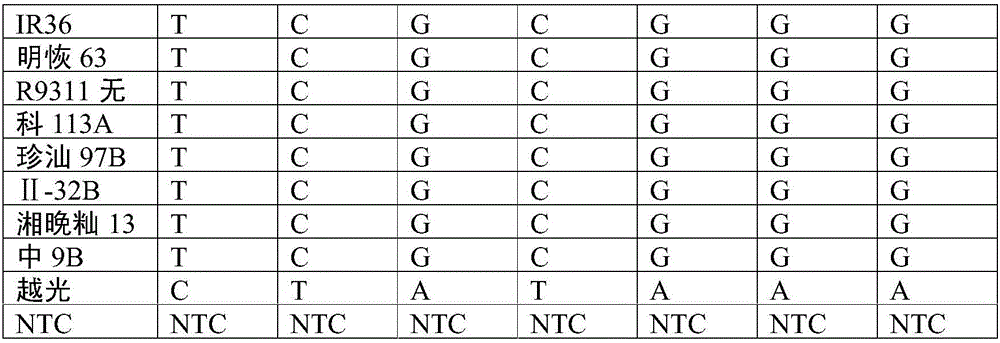
[0033] According to literature data, the rice OsHMA3 gene was mapped to the physical location on chromosome 7: 7405745-7409553 interval. The gene interval was expanded by 50kb to both sides, and SNP sites were extracted based on 3,000 rice resequencing data from the International Rice Institute, and selected according to the PIC value and whether there were other SNP sites in the 50 bp around the SNP site. The selected SNP sites were designed with primers using BatchPrimer3. For these SNP markers, KASP reaction verification was performed on two gene donor materials of rice varieties Nipponabre and Sasanishiki containing the OsHMA3 gene, two rice varieties of Chokokoku and Habataki that were determined not to have the function of the OsHMA3 gene, and 19 other high-cadmium and low-cadmium materials , to select SNP markers with good co-segregation and amplification effects with th...
Embodiment 2
[0045] Example 2 Application of SNP Molecular Marker of Rice Low Cadmium Accumulation Gene OsHMA3
[0046] 1. Extract the genomic DNA of the rice variety to be tested
[0047] 2. Amplify the nucleotide fragment containing the SNP site
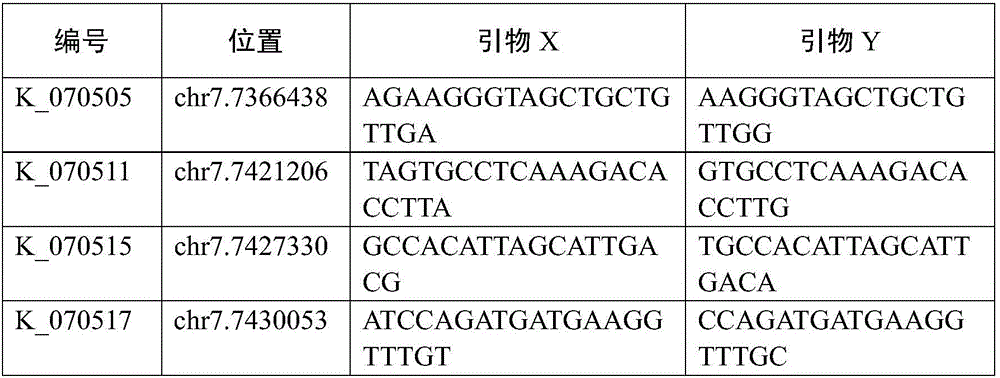
[0048] For the SNP site screened in Example 1, according to the primers of TagSNP-K_070520 (see Table 2), using genomic DNA as a template, amplify the nucleotide fragment where the SNP to be tested is located, as shown in SEQ ID NO.1 Show. The SNP site is located at 22bp of the PCR amplified fragment, where the base polymorphism is A or G.
[0049] KASP response tests were performed on the LGC SNPline genotyping platform. Add 20ng of DNA sample into the microwell reaction plate, add the KASP reaction mixture after drying, the reaction system is shown in Table 5.
[0050] Table 5 Reaction system for KASP detection
[0051] Final concentration Volume (μL) 100UM Primer C 0.42UM 0.0125 100UM Primer X 0.17UM 0.0050...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com