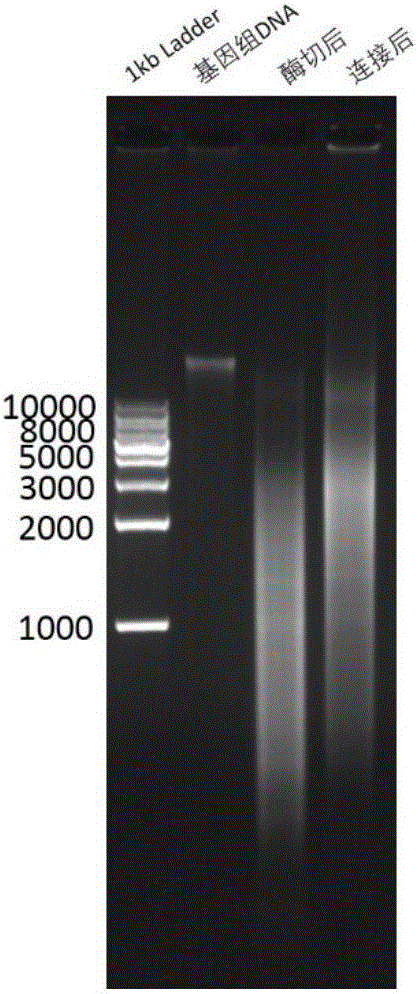
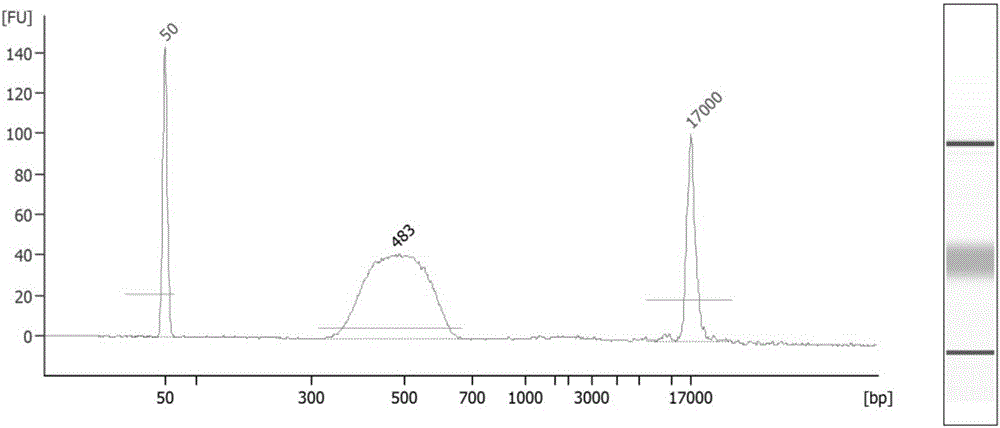
Method for capturing interacted DNA fragments in tissue nuclear genome
A technology for organizing cells and nuclei, which is applied in the field of molecular biology experiments, can solve the problems of unrealistic culture of cell lines, and achieve the effect of expanding the scope of use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The principles and features of the present invention are described below in conjunction with examples, which are only used to explain the present invention and are not intended to limit the scope of the present invention.
[0029] 1. Process animal tissue to obtain dispersed histiocytes
[0030] 1) Take 1g of fresh animal tissue sample, cut it into pieces as much as possible, put it on a 50ml centrifuge tube with a cell sieve (40μm), add 5mL room temperature 1×PBS to wash, and discard the PBS;
[0031] 2) Slowly and continuously add 10 mL room temperature 1×PBS, and squeeze the ground tissue sample through the cell sieve with a syringe plunger, and collect the filtrate in a 50 mL centrifuge tube;
[0032] 3) Centrifuge at 2000g RT for 5min, and discard the supernatant.
[0033] 2. Formaldehyde crosslinking
[0034] 1) Add 10 mL of formaldehyde with a final concentration of 1% prepared in 1×PBS, gently pipette and resuspend the cells, and shake at room temperature for ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com