Esophageal carcinoma prognosis marker and application thereof
A prognostic marker and technology for esophageal cancer, applied in the field of genetic engineering and oncology, to achieve the effect of improving accuracy and sequence conservation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
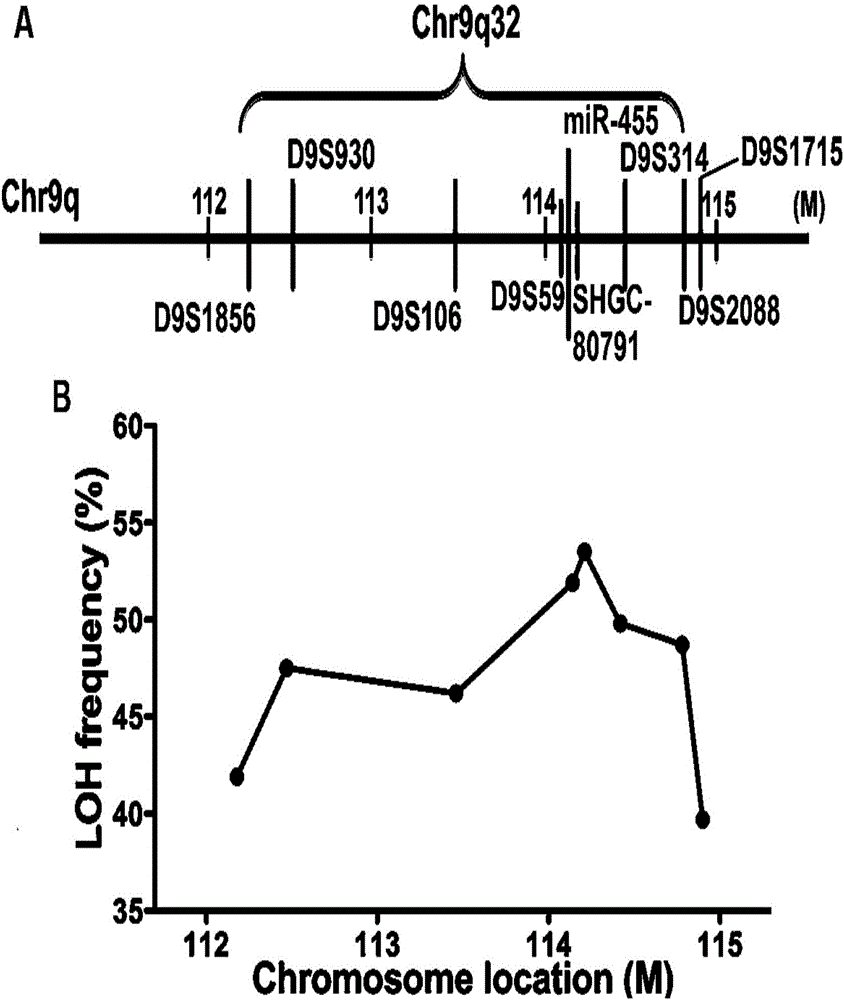
[0061] Example 1 Detection of loss of heterozygosity in the 9q32 region
[0062] The inventor collected 120 freshly resected esophageal cancer samples and 60 paired esophageal cancer samples at the General Hospital of the Chinese People's Liberation Army, and systematically collected the demographic and clinical data of these samples.
[0063] Genomic DNA was extracted from the above 60 paired esophageal cancer specimens (using QIAGEN’s Blood&Tissue Kit), the specific steps are:
[0064] 1. Weigh 20mg of tissue into a 1.5ml centrifuge tube, add 180μl Buffer ATL, then add 20μl proteinase K, vortex to mix, incubate at 56°C until the tissue is completely lysed, and vortex several times during the incubation.
[0065] 2. Add 200 μl Buffer AL, vortex to mix, and incubate at 56°C for 10 minutes.
[0066] 3. Add 200 μl of absolute ethanol and vortex to mix.
[0067] 4. Transfer all the above mixed solution into the DNeasy Mini spin column, centrifuge at 8000rpm for 1min, and disc...
Embodiment 2
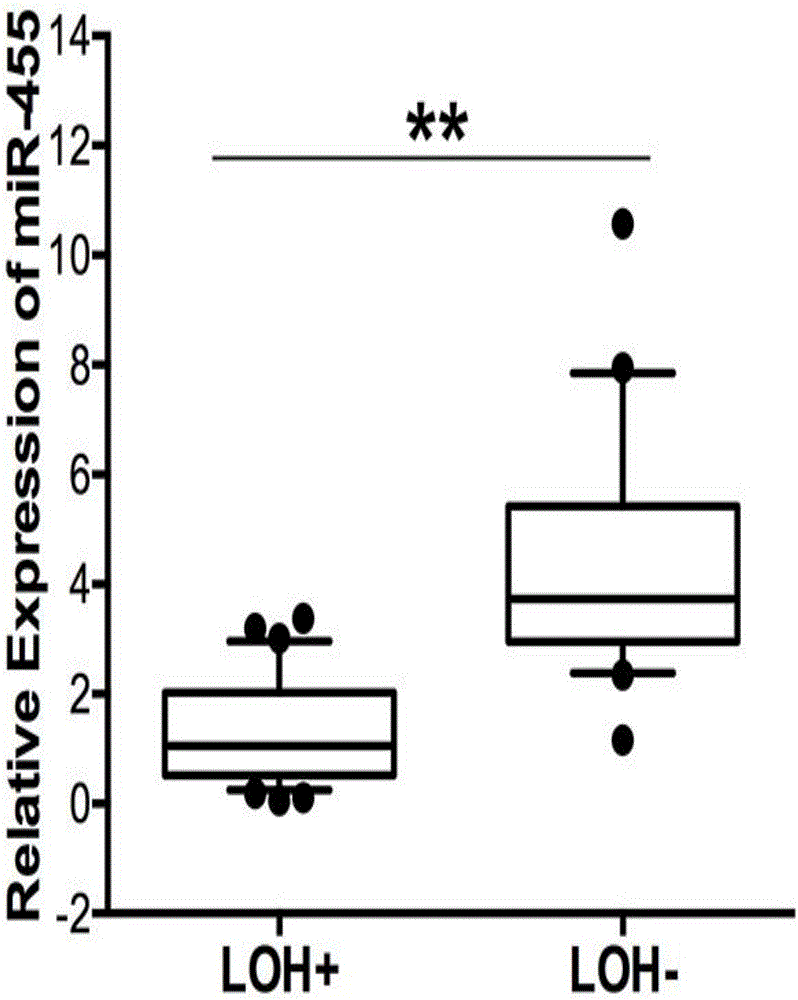
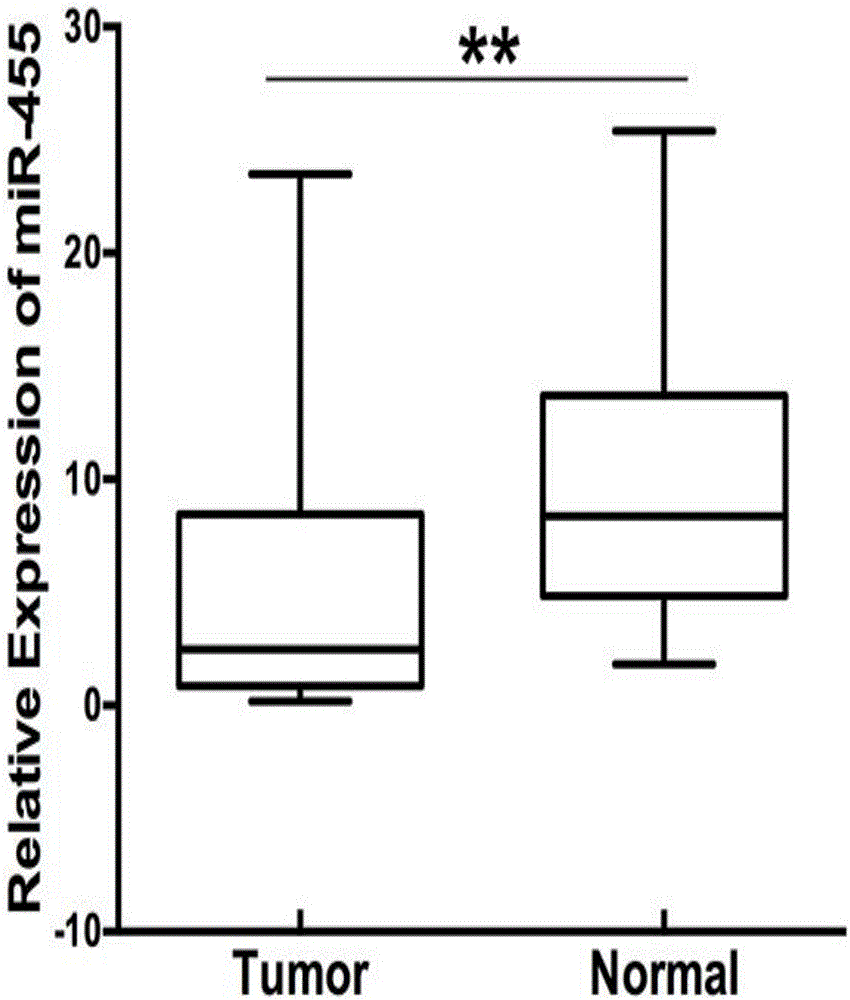
[0083] Example 2 Detection of miR-455 expression
[0084] 1. Total RNA was extracted from 120 esophageal cancer samples and 60 paired esophageal cancer samples. The specific method is as follows:
[0085] 2. Take 20mg of each sample, add 1ml TriZol reagent, grind, centrifuge at 12000g for 2min, take the upper layer liquid into a new 1.5ml centrifuge tube.
[0086] 3. Incubate on ice for 15min, add 200μl chloroform, vortex for 30s, incubate for 5min, centrifuge at 12000g, 4°C for 10min.
[0087] 4. Take the upper aqueous phase to a new 1.5ml centrifuge tube, add 500μl isopropanol, incubate on ice for 15min, centrifuge at 12000g, 4°C for 15min, discard the supernatant.
[0088] 5. Add 1ml of 75% ethanol, vortex for 10s, centrifuge at 6000g for 5min at 4°C, discard the supernatant, and dry in the air.
[0089] 6. Add appropriate amount of RNase-free water to dissolve.
[0090] RNA with poly(U) tails (New England Biolabs)
[0091] 1. Take 10ng of total RNA, UTP, poly(U) polyme...
Embodiment 3
[0107] Embodiment 3 Implementation is used to assist in judging the miRNA kit making of the prognosis of patients with esophageal cancer
[0108] (1) The production and operation process of the miRNA kit is based on technologies such as total RNA plus poly(U) tail, RT-PCR and Real-time PCR.
[0109] Kit includes:
[0110] Primers (including reverse transcription universal primer SL-poly(A): GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAAAAAAAAAAAAAAAVN;
[0111] miR-455 upstream primer: GAACTGCAGTCCATGGGCATA;
[0112] Internal reference U6 upstream primer: CTCGCTTCGGCAGCACA;
[0113] Universal downstream primer: GCAGGGTCCGAGGTATTC).
[0114] Reagents required for PCR reaction, including: RNA isolation solution, PCR reaction solution, buffer, poly(U) polymerase, reverse transcriptase, universal reverse transcription primer (poly(A)
[0115] RNA separation solution; the RNA separation solution is composed of Tween 20, trishydroxymethylaminomethane, ethylenediaminetetraacet...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com