Forecasting method for forecasting DNA-mutation-influenced protein-protein interaction based on multivariate data
A technology of protein interaction and multivariate data, applied in proteomics, electronic digital data processing, special data processing applications, etc., to achieve the effect of improving robustness and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
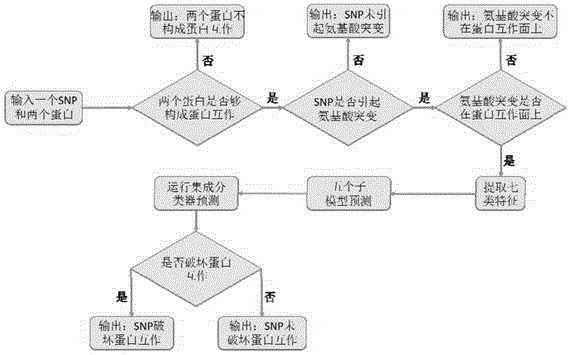
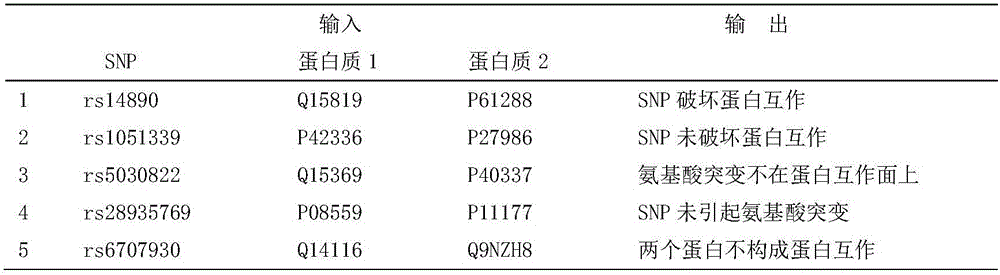
[0060] Step 1. Organize all non-redundant protein interaction pairs contained in the five protein interaction databases of HPRD, BioGrid, IntAct, MINT and DIP, a total of 233,461 pairs, which can be used to determine whether there is protein interaction between proteins.
[0061] Step 2. Combining the 161,456,298 dbSNP data provided by NCBI, use the software Polyphen2, SIFT, and MutationAsseso to calculate the database of amino acid mutations on protein sequences caused by SNPs, with a total of 33,306 records, which can be used to determine whether SNPs cause amino acid mutations on proteins.
[0062] Step 3. Based on the protein interaction structure information provided in the PDB database, a total of 260,182 pieces of protein interaction surface information were sorted out. Using the database in Step 2, it was possible to determine whether the amino acid mutation caused by the SNP occurred on the protein interaction surface.
[0063] Step 4. Apply the SNP-protein interaction...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com