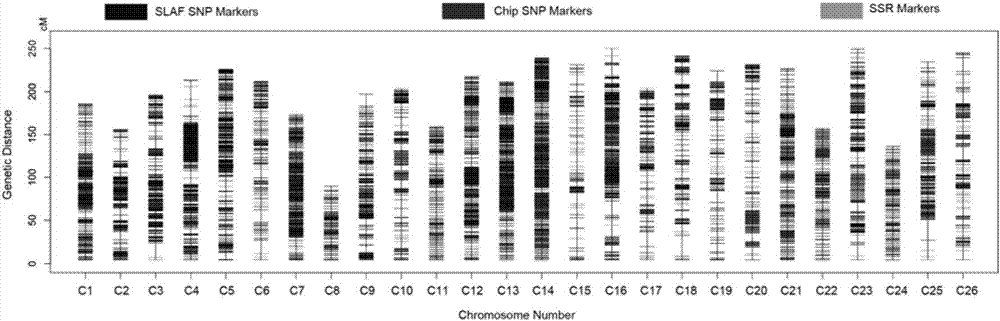
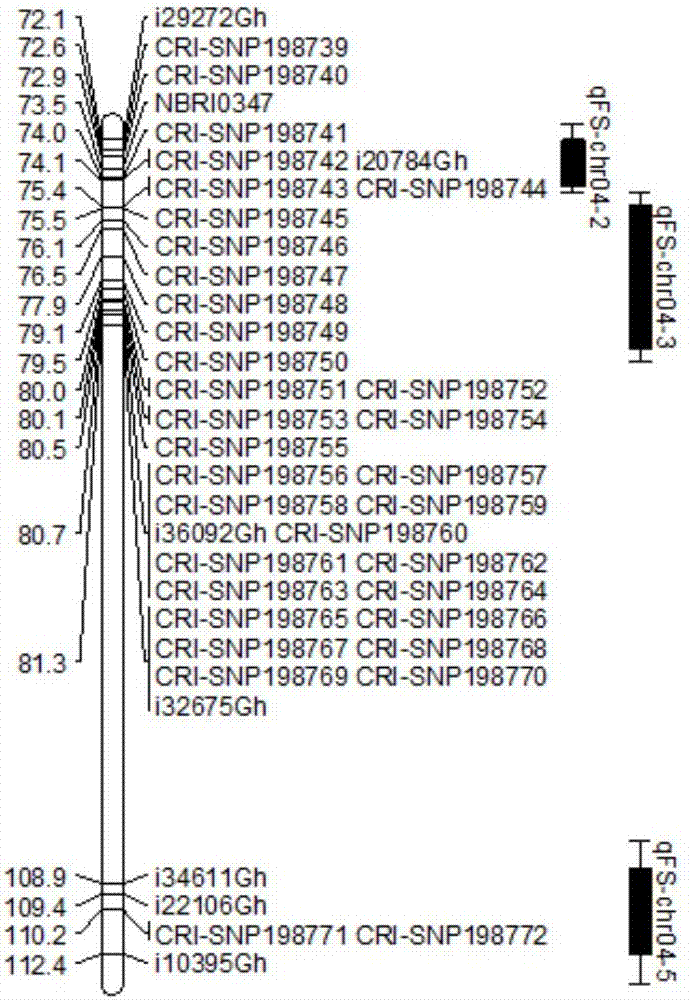
Upland cotton No. 4 chromosome and SNP molecular markers associated with fiber strength
A fiber strength and molecular marker technology, which can be used in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of complex genetic background, less cotton application, lack of reliability and stability, etc., and improve fiber quality. , the effect of improving breeding efficiency and shortening breeding cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0022] The present invention will be further clarified through the detailed description of specific embodiments below, but it is not intended to limit the present invention, but only for illustration.
[0023] (1) Recombinant inbred line F 6:8 the acquisition
[0024] For the field planting and DNA extraction from 2007 to 2008, please refer to the patent application publication number: CN 101613761A, the title of invention: the patent application document of the SSR marker linked to the main effect gene of cotton fiber strength. In 2009, the parents and F 6:10group. Anyang and Quzhou adopt a single-row area with a length of 5 meters and a row spacing of 0.8m and (0.8+0.5)m respectively, with 20 plants in each row; Xinjiang adopts a 6-row area with a length of 2 meters and 15 plants in each row. In 2010, the parents and F 6:11 For groups, Anyang and Zhengzhou adopt single-track areas with a row length of 5m and a row spacing of 0.8m; Gaoyi adopts a single-row area with a ro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com