Method for constructing stable cell strain secreting expression protein
A technology of secretion expression and construction method, which is applied to genetically modified cells, cells modified by introducing foreign genetic material, animal cells, etc., which can solve the problems of heavy workload and difficult purification, and achieve low cost and low reagent cost , the effect of less impurity protein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Construction of pCDH-S-His recombinant lentiviral vector
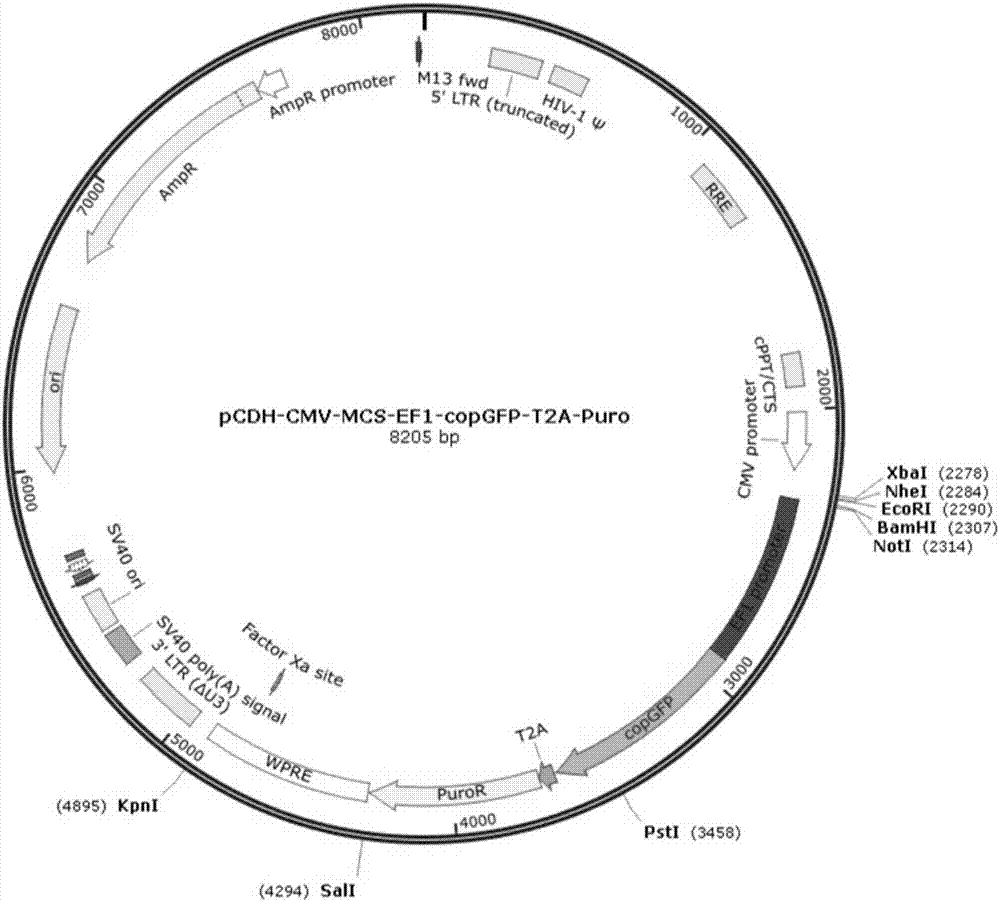
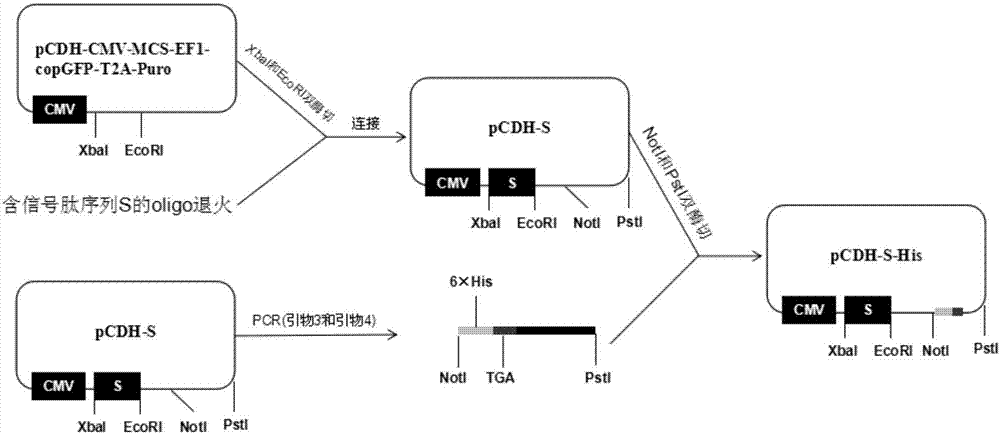
[0039] Plasmid pCDH-CMV-MCS-EF1-copGFP-T2A-Puro( figure 1 ) to perform double digestion with XbaI and EcoRI to recover the vector backbone. Such as figure 2 as shown,
[0040] Design and synthesize 2 oligonucleotide chains with the following sequences:
[0041] Forward: (5'→3') with XbaI sticky ends
[0042] CTAGAATGGAGACAGACACACTCCTGCTGTGGGTGCTGCTGCTCTGGGTCCCAGGTCCCACTGGCGACGGG (SEQ ID NO. 7);
[0043] Reverse: (5'→3') with EcoRI cohesive ends
[0044] AATTCCCGTCGCCAGTGGAGCCTGGGACCCAGAGCAGCAGCACCCACAGCAGGAGTGTGTCTGTCTCCATA (SEQ ID NO. 8).
[0045] Anneal the above two oligonucleotide chains, and connect them with the backbone of the carrier at 16°C with ligase, transform into E. coli overnight for 1 hour, and pick a single clone colony for overnight culture the next day, extract the plasmid and verify it by sequencing, and finally obtain the recombinant Vector pCDH-S.
[0046] The recombinant...
Embodiment 2
[0054] Example 2 Construction of pCDH-S-PEDF-His recombinant plasmid
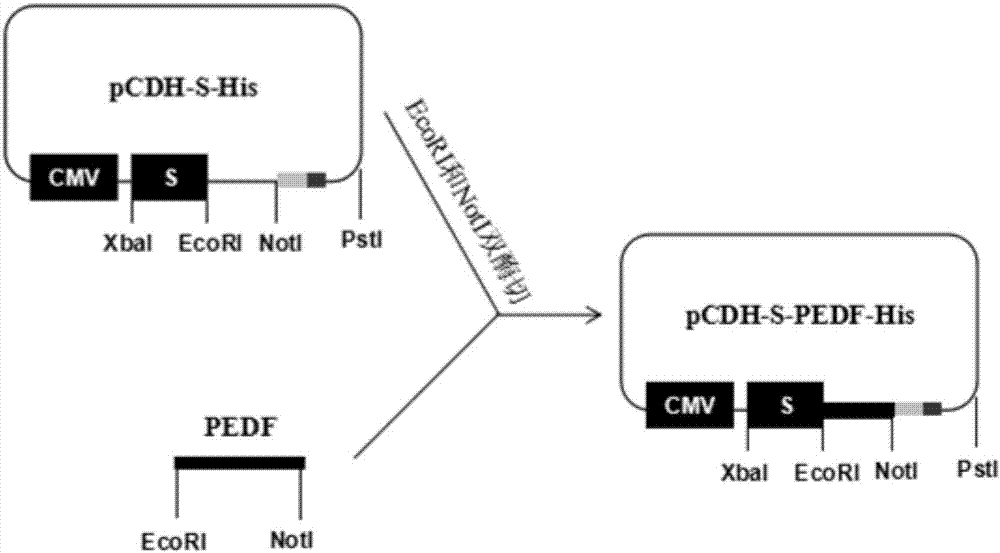
[0055] Such as image 3 as shown,
[0056] Using the recombinant lentiviral vector pCDH-S-His as the backbone, the backbone part was recovered by double digestion with EcoRI and NotI.
[0057] Primers were designed to amplify the PEDF gene, and the primer sequences were as follows:
[0058] Upstream primers:
[0059] 5'-TTTGAATTCTATGCAGGCCCTGGTGCTA-3' (SEQ ID NO. 1);
[0060] Downstream primers:
[0061] 5'-TTTGCGGCCGCGGGGCCCCTGGGGTCCAGAA-3' (SEQ ID NO. 2);
[0062] Using human cDNA as a template, the PEDF gene was obtained by PCR reaction. The amplification program was pre-denaturation at 95°C for 4 minutes, denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute and 20 seconds, 35 cycles, and finally extension at 72°C for 10 minutes.
[0063] After the PCR product was purified, it was digested with EcoRI and NotI, recovered by running the gel, and then ...
Embodiment 3
[0064] Example 3 Construction of pCDH-S-PLGF-His recombinant plasmid
[0065] Such as Figure 4 as shown,
[0066] Using the recombinant lentiviral vector pCDH-S-His as the backbone, the backbone part was recovered by double digestion with EcoRI and NotI.
[0067] Design primers to amplify the PLGF gene, and the primer sequences are as follows:
[0068] Upstream primers:
[0069] 5'-TTTGAATTCCCTGCCTGCTGTGCCCCCCCA-3' (SEQ ID NO.3);
[0070] Downstream primers:
[0071] 5'-TTTGCGGCCGCGCCTCCGGGGAACAGCAT-3' (SEQ ID NO.4);
[0072] Using human cDNA as a template, the PLGF gene was obtained by PCR reaction. The amplification program was pre-denaturation at 95°C for 4 minutes, denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute and 20 seconds, 35 cycles, and finally extension at 72°C for 10 minutes.
[0073] After the PCR product was purified, it was digested with EcoRI and NotI, recovered by running the gel, and then ligated w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com