Auxiliary genome assembly method based on artificial meiosis
A genome assembly and meiosis technology, applied in the field of genomics, can solve the problems of requiring library expansion, time-consuming and labor-intensive, large-scale application limitations, etc., and achieve the effects of fine and reliable physical map, simple experimental operation, and simple operation.
Active Publication Date: 2017-08-18
OCEAN UNIV OF CHINA
View PDF2 Cites 6 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment
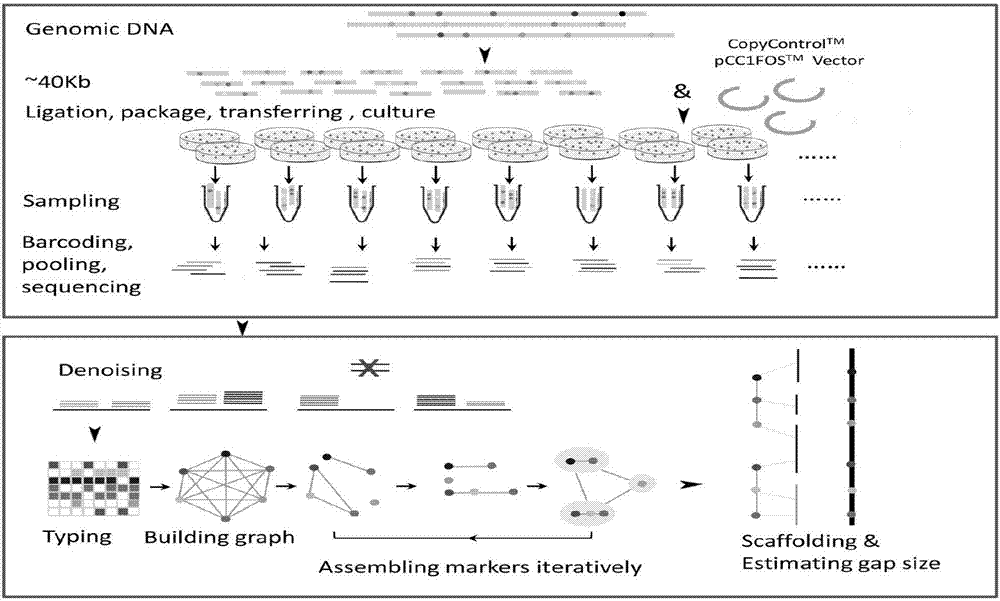
[0033] Taking Arabidopsis thaliana as an example, the assembly process of the Arabidopsis genome is as follows: figure 1 shown.
[0034] (1) Experimental library construction
[0035] 1) Extraction of Arabidopsis genomic DNA
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention discloses an auxiliary genome assembly method based on artificial meiosis. A genome in a form of clone library is equally divided so as to establish random artificial meiosis samples, and the random artificial samples are treated through Hpall transmethylase and FspEi methyl-modified dependent-type incision enzyme to form and analyze typing markers with high densities; and then sorting information of the typing markers is obtained; therefore, further assembly of scaffold or direct cascade assembly of Pacbio sequencing reads are achieved. Based on an assembly strategy of artificial meiosis, the auxiliary genome assembly method has the advantages of being easy in experiment operation, short in period, low in cost and capable of splicing the genome with high coverage and accuracy, and owns a wide application prospect in species with relatively complicated and highly heterozygous genomes.
Description
technical field [0001] The invention relates to the field of genomics, in particular to an auxiliary genome assembly method based on artificial meiosis. [0002] technical background [0003] Genomic DNA sequence is the main carrier of the genetic information of organisms. With the help of whole genome sequencing and the interpretation of sequence information, the growth and development principles of many important species can be revealed at the molecular level, and the genetic differences among individuals can also be explored at the population level. It is of great significance to explore and understand the nature of life and other basic biological science research, the prevention and treatment of important human genetic diseases, and applied research such as animal and plant genetic breeding. [0004] Whole-genome sequencing technology has evolved from the first-generation dideoxy chain termination method to the second-generation sequencing method of sequencing-by-synthesi...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12N15/10
Inventor 王师包振民窦锦壮窦怀乾张玲玲吕佳
Owner OCEAN UNIV OF CHINA
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com