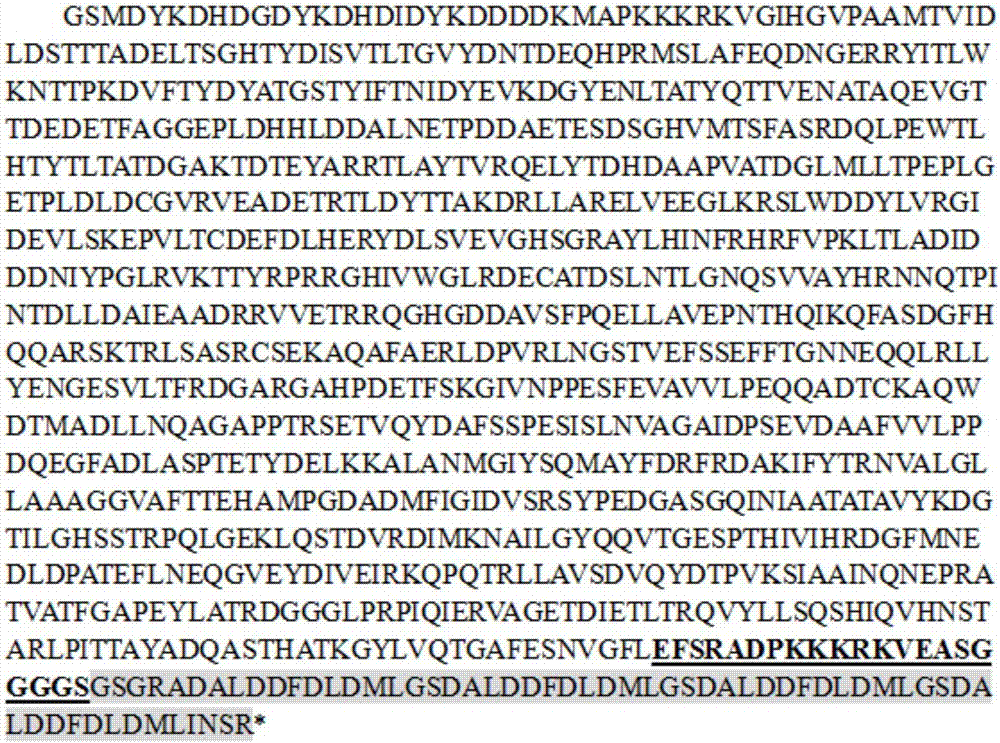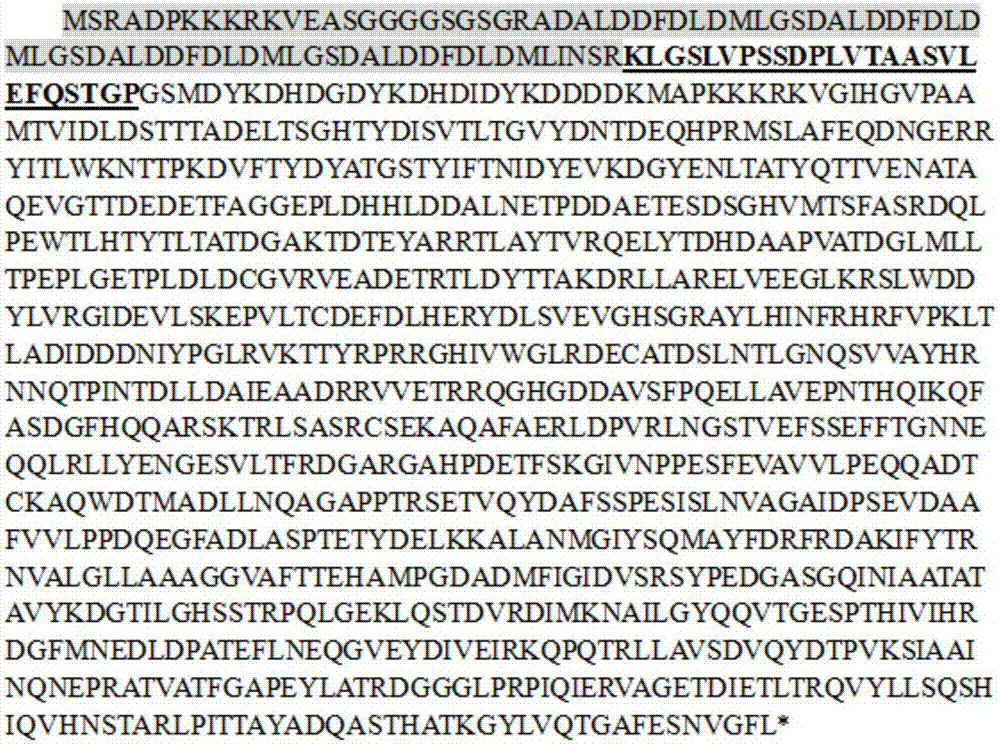Method for targetedly activating gene transcription on basis of natronobacterium gregoryi Argonaute
A technology of gene editing and endogenous genes, applied in genetic engineering, chemical instruments and methods, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
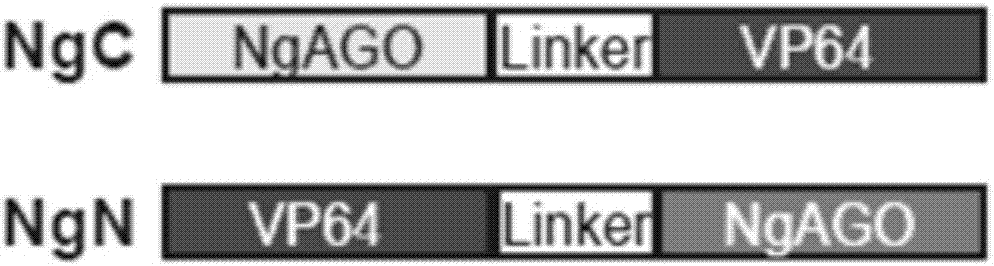
[0040] In the following examples, taking the Nanog gene as an example, the research on the activation of intracellular genes by using NgC (fused VP64 to the C-terminal of NgAGO) and NgN (fused VP64 to the N-terminal of NgAGO) was carried out. Example 1 Construction of NgAGO-VP64 fusion protein
[0041] 1. Fusion expression of VP64 and NgAGO
[0042] In order to prevent the influence of the relative position of the protein on the protein activity, VP64 was fused to the C-terminal (NgC) and N-terminal (NgN) of NgAGO respectively, and the protein sequences were shown in SEQ ID NO.1 and SEQ ID NO.2 respectively (gene structure Schematic such as figure 1 As shown, the sequence composition is as follows figure 2 and image 3 shown).
[0043] 2. After the protein sequence is optimized for humanized codons, it is cloned into the mammalian expression vector pCDNA3.1 respectively. After the plasmid was verified to be correct by sequencing, the plasmid was extracted and used for subs...
Embodiment 2
[0044] Example 2 Guided DNA Synthesis and Phosphorylation
[0045] 1. Design the guide DNA sequence, as shown in Table 1:
[0046] Table 1
[0047]
[0048] The guide DNA (gDNA) is 24bp in length and dissolved to 100μM with 1XT4PNK buffer (NEB) after synthesis. Phosphorylate with 5 units of T4PNK per 1nmolgDNA, and incubate overnight at 37°C. Then store at -20°C for later use.
Embodiment 3
[0049] Example 3 NgAGO genome editing tool composed of NgAGO-VP64 fusion protein and guide DNA and its application
[0050] 1. An NgAGO genome editing tool based on Halophilus grisii Argonaute targeted activation gene transcription, comprising the NgAGO-VP64 fusion protein described in Example 1 and the guide DNA described in Example 2.
[0051] 2. NgAGO genome editing tool activates gene transcription experiment
[0052] (1) Cell culture and transfection
[0053] The day before transfection, the cells were subcultured so that the cells were in the exponential growth phase at the time of transfection. Taking Hela cells as an example, 100,000 cells can be plated in each well of a 12-well plate. One hour before transfection, the cell culture medium was changed to serum-free medium and incubated for 1 hour. Transfection was performed using Lipofactamine2000 transfection reagent.
[0054] Taking the amount per well in a 12-well plate as an example, dilute 1 μg fusion protein p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com