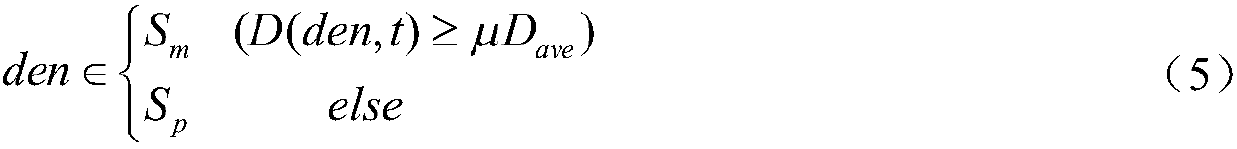Cancer subtype precise discovery and evolution analysis method based on data stream clustering
A technology of data flow clustering and analysis method, applied in the field of cancer subtype discovery and evolution analysis, can solve problems such as hindering the production of clustering results, and achieve the effect of high precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] The present invention will be further described below in conjunction with the accompanying drawings.
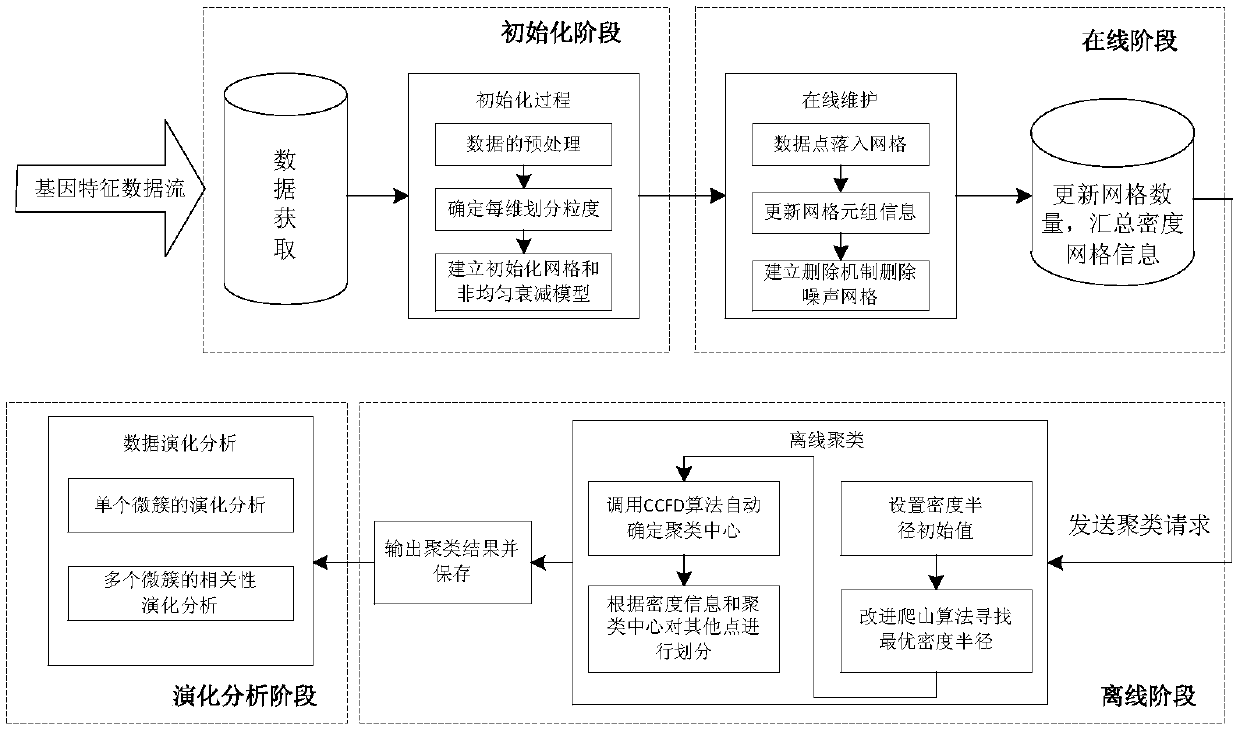
[0039] refer to figure 1 and figure 2 , a method for precise discovery and evolution analysis of cancer subtypes based on data stream clustering, including the following steps:
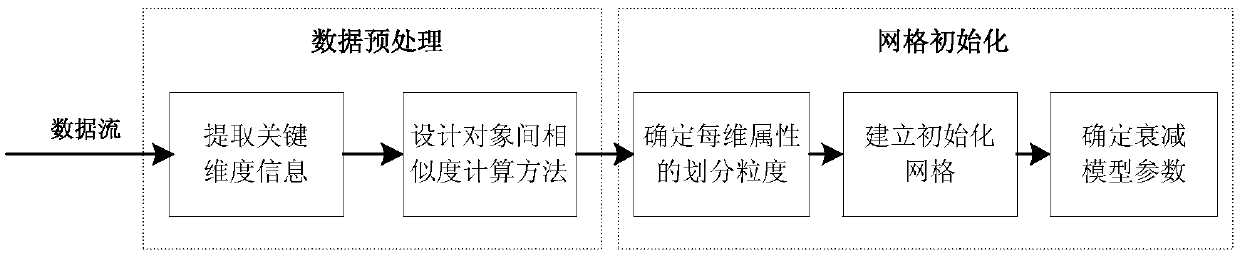
[0040] (a) Initialization of the gene expression data stream. Gene data flow data preprocessing operation: analyze the dimensional information of the data flow, and determine the calculation method of similarity distance; establish the grid unit of the genetic data flow object, and put the data into the grid by window to realize initialization; construct non-uniform Attenuation model, which determines the non-uniform attenuation parameters of the data flow in the online process and the update method of the grid density information.
[0041] (b) Online real-time clustering of gene expression data streams. In order to ensure real-time clustering requirements, each arriving data point is put...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com