Method for detecting microRNA based on triple helix DNA structure
A structure detection and triple helix technology, applied in the field of fluorescent microRNA detection, can solve the problems of time-consuming and large number of samples, and achieve the effect of fast response and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
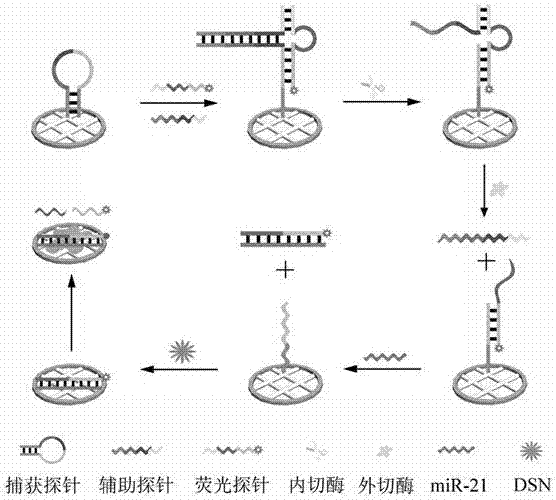
[0028] 1. a method for detecting microRNA based on triple helix DNA structure, is characterized in that comprising the following steps:
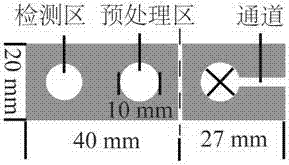
[0029] 1.1 Use Adobe illustrator CS4 to design the hydrophobic wax batch printing pattern of the microfluidic paper chip;
[0030] 1.2 Cut the paper into a commonly used A4 size paper, print the hydrophobic wax batch printing pattern designed in step (1.1) onto the A4 paper, and then place the A4 paper with the wax pattern in a flat heater or an oven at 150 ºC heating for 2 min; the wax melts and soaks through the entire thickness of the paper to form a hydrophobic wall;
[0031] 1.3 Probe pre-denaturation
[0032] First, capture probes, auxiliary probes and fluorescent probes were incubated at 95 ºC for 5 min, and the three solutions were naturally cooled at room temperature for 30 min;
[0033] 1.4 Construction of triple helix DNA structure
[0034] Immobilize 80 nM sulfhydryl-labeled capture probes on the surface of the microfluidic pa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com