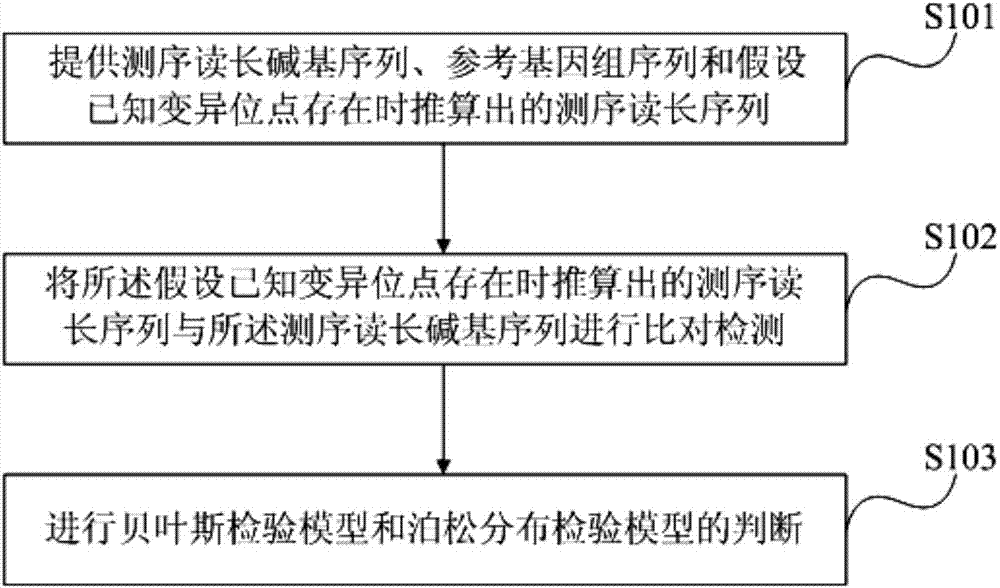
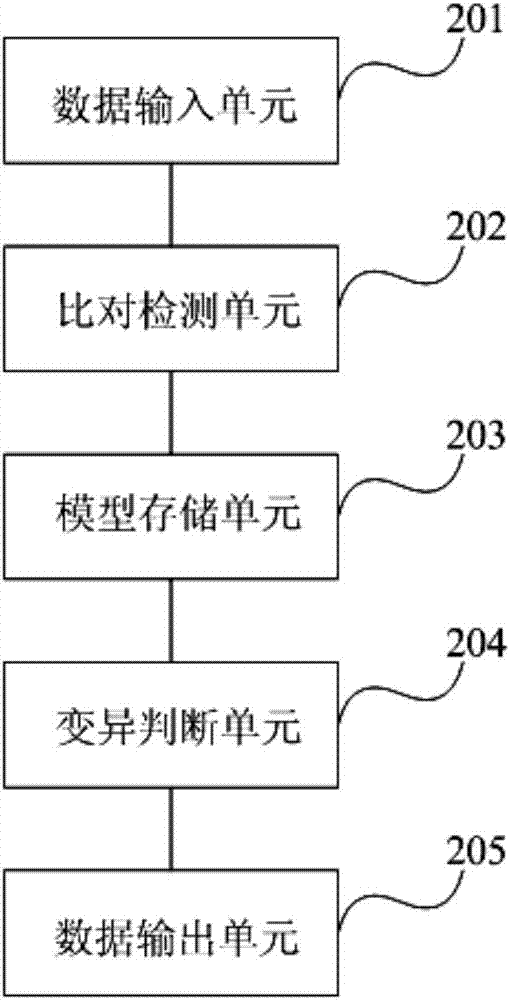
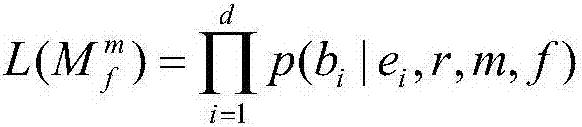Method and device for known variation detection based on Bayes testing and Poisson's distribution testing
A technology of Poisson distribution and variation, applied in the field of known variation detection, can solve the problems of lower comparison quality, inability to detect multiple genes and multiple sites at the same time, and inability to detect positional mutations, etc., to reduce false positives and avoid The effect of comparison quality degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] The specific parameters used by the detection method of the present embodiment are set to:
[0079] Sample DNA: FFPE tissue sample from a female patient with upper left lung adenocarcinoma;
[0080] Sample processing and sequencing off-machine data: target region capture and BGISEQ-100 platform sequencing;
[0081] Sequencing off-machine data preprocessing and quality control: The effective data off-machine sequencing is compared to the reference genome through tmap, samtools sorting, BamDuplicates deduplication, samtools index indexing, QC quality control, and qualified files are followed by subsequent mutation detection steps ;
[0082] Reference Genome: the human genome reference sequence of version 37.3 (hg19; NCBI Build 37.3) in the NCBI database;
[0083] Sequencing read length calculated assuming that known variant sites exist: based on the wild type and variant column data in Table 1 and the reference genome sequence;
[0084] Bayesian test model: (L(M 0 )...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com