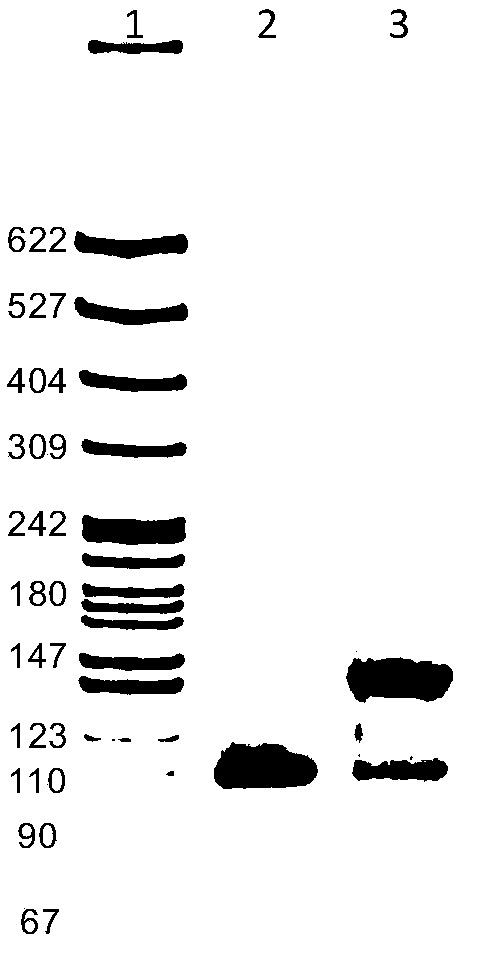
DNA (desoxyribonucleic acid) fluorescent hydrogel and preparation method thereof
A deoxyribonucleic acid and hydrogel technology, applied in medical science, DNA/RNA fragments, bandages, etc., can solve the problems of poor bioabsorbability and degradability, and achieve the effect of reducing wound pain, soft materials, and simple process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] A method for preparing circular DNA with primary primers, comprising the steps of:
[0046] The specific sequences of the phosphorylated linear single-stranded DNA at the 5' end of the circular DNA with the primary primer, the primary primer, and the ssDNA added to the synthetic deoxyribonucleic acid fluorescent hydrogel are shown in Table 1.
[0047] Table 1. DNA sequences
[0048]
[0049] Wherein the nucleotide sequence of the third part in the ssDNA fragment is: 5'-ATCCTCCCACCGGGCCTCCCAC-3'SEQID NO.10
[0050] (1) Synthesis of circular DNA with primary primers:
[0051] a. DNA sample pretreatment:
[0052] Use a mini centrifuge to centrifuge the phosphorylated linear single-stranded DNA at the 5' end, the primary primer and ssDNA respectively for 1 min, throw them to the bottom of the centrifuge tube, then dissolve them in ultrapure water to 100 μM, and shake them in a constant temperature shaker for 4 hours , the condition is 30°C, 1000rpm.
[0053] b. Mix t...
Embodiment 2
[0074] A preparation method for deoxyribonucleic acid fluorescent hydrogel, comprising the steps of:
[0075] (1) Perform rolling circle amplification to obtain DNA hydrogel:
[0076] a. add the circular DNA (prepared in Example 1) with the primary primer with a final concentration of 50nmol / L according to the amount in Table 5, the phi 29 DNA polymerase with a final concentration of 0.05U / μl, and the phi 29 DNA polymerase with a final concentration of 1mmol / L dNTPs and 10×phi 29 DNA polymerase buffer and 100×BSA, NaCl solution with a final concentration of 16mmol / L, the balance is ultrapure water, the reaction condition is 200rpm shaking for 100min, and the rolling circle amplification reaction is performed to obtain long single-strand DNA.
[0077] table 5
[0078]
[0079] b. Take 100 μl of the reaction solution obtained in step a, add 2 μl of 100 μmol / L ssDNA aqueous solution, and shake at 200 rpm for 100 minutes.
[0080] The ssDNA sequence consists of three parts, ...
Embodiment 3
[0089] A method for preparing circular DNA with primary primers, comprising the steps of:
[0090] The specific sequences of the phosphorylated linear single-stranded DNA at the 5' end of the circular DNA with the primary primer, the primary primer, and the ssDNA added to the synthetic deoxyribonucleic acid fluorescent hydrogel are shown in Table 6.
[0091] Table 6. Deoxyribonucleic acid sequences
[0092]
[0093] Wherein the nucleotide sequence of the third part in the ssDNA fragment is preferred: 5'-CCCCCCCCCC-3'SEQ ID NO.11
[0094] (1) Synthesis of circular DNA with primary primers:
[0095] a. DNA sample pretreatment:
[0096] Use a mini centrifuge to centrifuge the phosphorylated linear single-stranded DNA at the 5' end, the primary primer and ssDNA respectively for 1 min, throw them to the bottom of the centrifuge tube, then dissolve them in ultrapure water to 100 μM, and shake them in a constant temperature shaker for 4 hours , the condition is 30°C, 1000rpm. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com