A kind of amplification primer, sequencing primer, kit and method for detecting hepatitis C virus genotyping
A technology for hepatitis C virus and genotyping, which is applied in the field of kits, sequencing primers, and amplification primers for detecting hepatitis C virus genotyping, can solve the problem of high price, not easy operation, low sensitivity and low specificity and other problems, to achieve the effect of high sensitivity, easy detection and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1 detects the primers of hepatitis C virus genotyping
[0036] Primers for detection of hepatitis C virus genotyping based on Sanger sequencing technology, including amplification primers and sequencing primers.
[0037]Design PCR primers and corresponding sequencing primers for the core / E1 region of HCV genomic RNA, use primer design software, such as Primer Premier 5, to select a specific sequence from the core / E1 region sequence of HCV genomic RNA, and then base , to design specific primers. The primer sequences are SEQ ID No: 1-2, respectively.
[0038] name serial number Sequence 5'-3' CE-F SEQ ID NO.1 CGCAATGTGCGTAACGTCAACGATAC CE-R SEQ ID NO.2 GTAGGCGACGACTTCTTCATCA
[0039] The design of the sequencing primer is similar to that of the specific primer used to amplify the viral RNA in the sample, but only one sequencing primer is required, and the primer sequence is SEQ ID No:3.
[0040] name serial num...
Embodiment 2
[0041] Embodiment 2 detects the kit of hepatitis C virus genotyping
[0042] The test kit of the detection hepatitis C virus genotyping of the present embodiment comprises the following table components:`
[0043]
[0044] 1. PCR detection reagents, including amplification primers (with base sequences shown in SEQ ID No: 1 and SEQ ID No: 2), Tris hydrochloride Buffer, hot start Taq enzyme (hot start Taq enzyme adopts chemical modification, through Heat shock at 95°C for 15 minutes, Taq enzyme can play a role, hot start Taq enzyme can reduce the formation of primer dimers and increase reaction specificity), reverse transcriptase.
[0045] 2. PCR sequencing reagents, including sequencing primers (with the base sequence shown in SEQ ID No: 3), sequencing dye (BigDye), sequencing reaction solution (BigDye Sequencing Buffer), high-purity deionized formamide (Hi-DiFormamide )
[0046] 3. Quality control products, including negative quality control products and HCV typing positi...
Embodiment 3
[0048] Embodiment 3 detects the method for hepatitis C virus genotyping
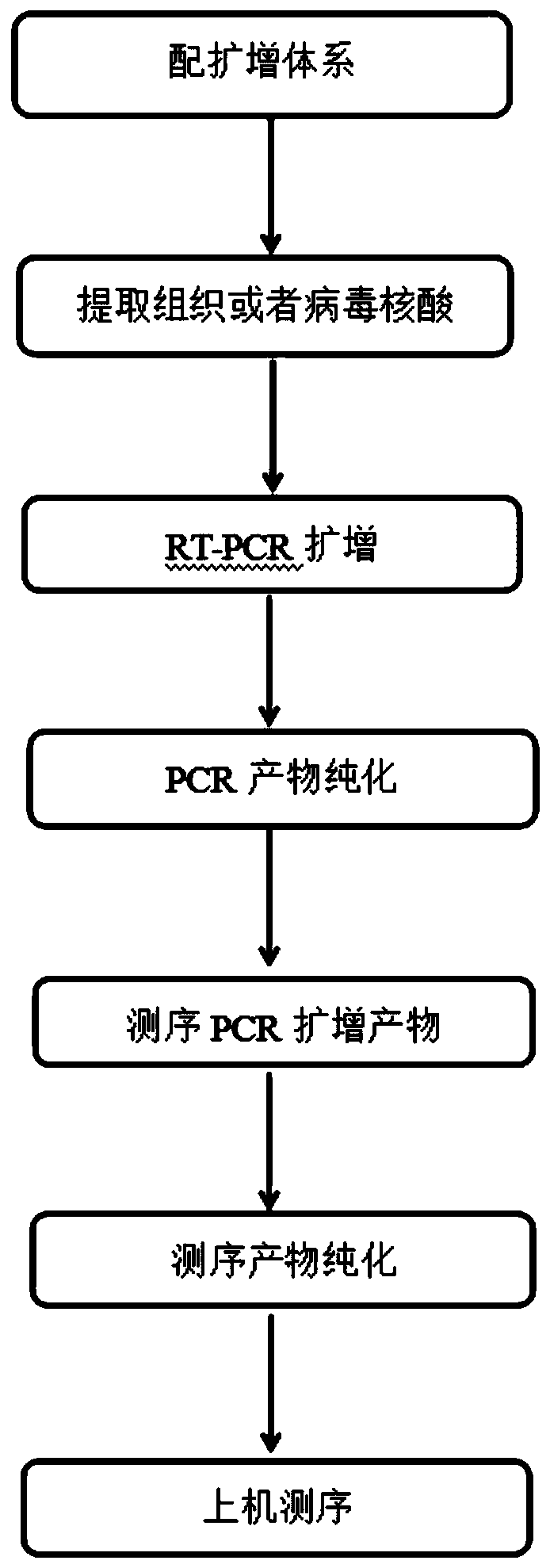
[0049] see figure 1 , is the operation flowchart of the method for detecting hepatitis C virus genotyping of the present invention; The method for detecting hepatitis C virus genotyping of the present embodiment comprises the following steps:
[0050] 1. Take out the HCV RT-PCR reaction solution and HCV RT-PCR enzyme system from the kit, thaw at room temperature, shake and mix well, and centrifuge at 8,000rpm for a few seconds before use.
[0051] Take N (N = number of samples to be tested + HCV typing positive quality control + negative quality control) reaction tubes,
[0052] The HCV single RT-PCR amplification system is prepared in the following table
[0053] components HCV RT-PCR reaction solution HCV RT-PCR Reaction Solution Enzyme System total capacity Dosage 27μl 3μl 30μl
[0054] Mix the components thoroughly, and then centrifuge for a short time to centrifuge ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com