A droplet-based method and apparatus for composite single-cell nucleic acid analysis
A cell and droplet technology, applied in biochemical equipment and methods, recording carriers used by machines, DNA preparation, etc., can solve the problem of high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
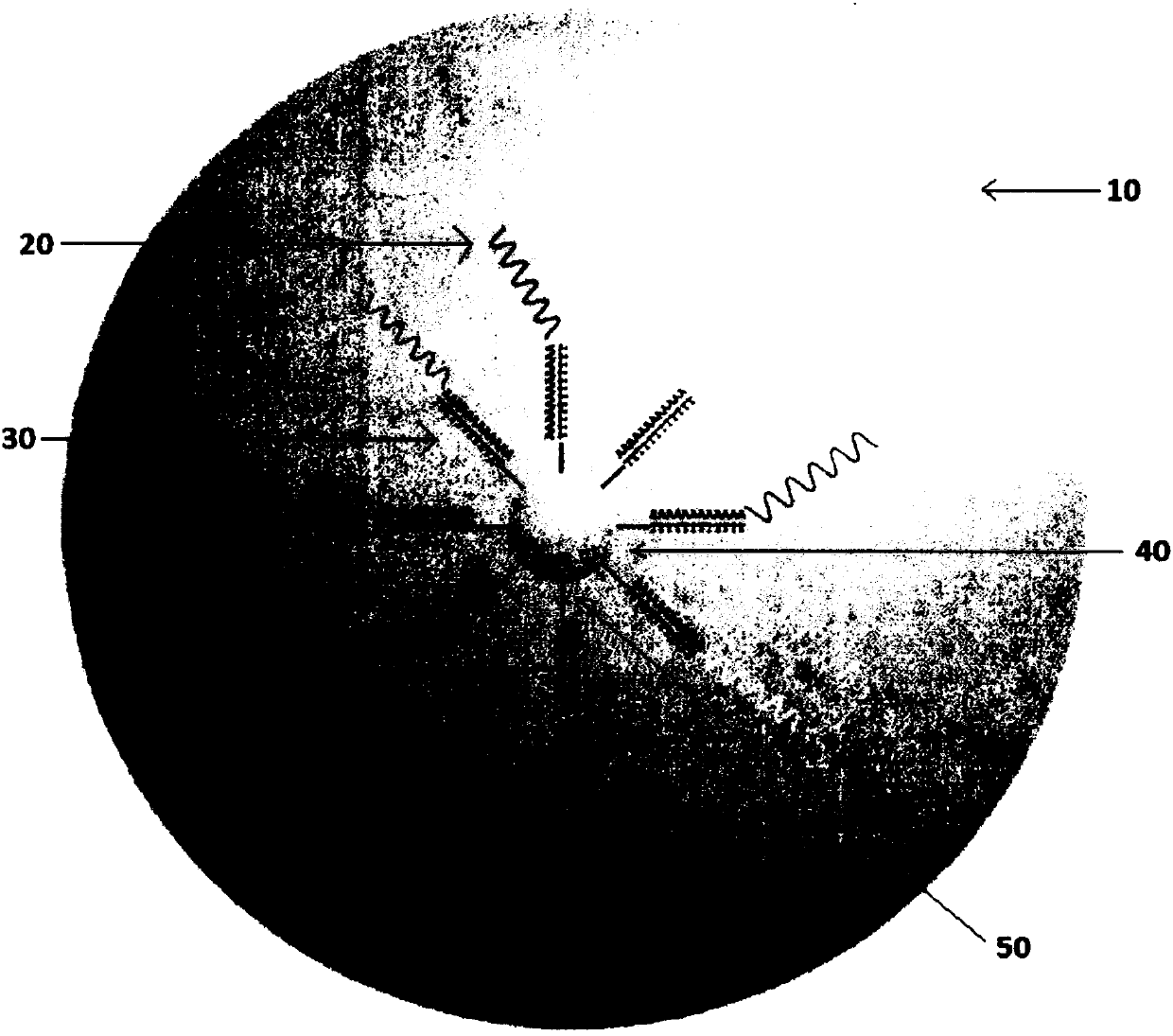
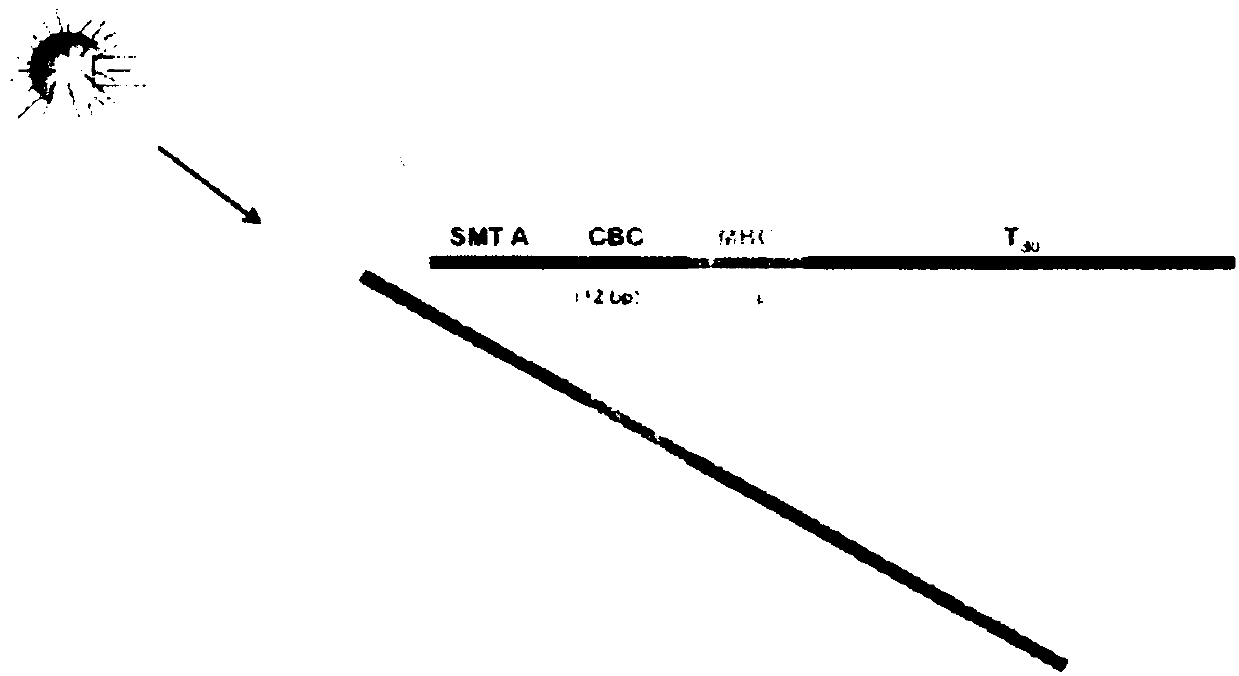
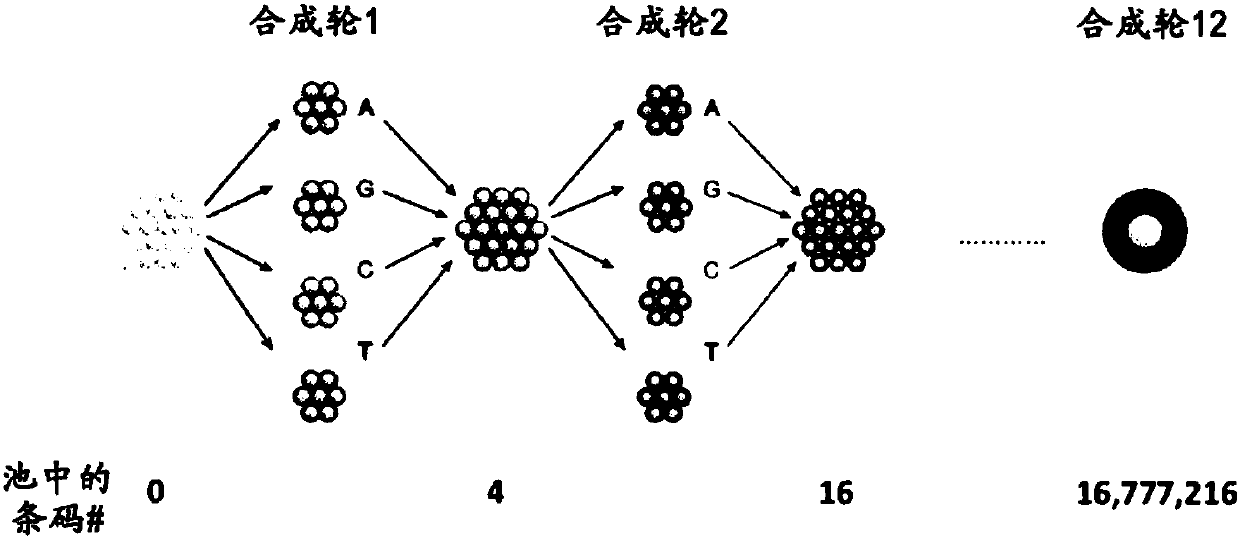
[0162] In this protocol, uniquely barcoded beads were synthesized for use as primers for reverse transcription. Beads are first prepared with immobilized sequences synthesized on the surface ( Figure 2A A) starts with the SMT in A), which serves as a priming site for downstream PCR. Next, the beads were split and pooled a total of 12 times into four identical reaction vessels to generate 4^12 unique barcode sequences unique to each bead ( Figure 2B ). This 12bp region was used as the cell barcode as it was specific for each bead. Next, the beads are all pooled together for 8 rounds of degenerate synthesis using all four bases; this 8bp region is the "molecular barcode" and will uniquely tag each mRNA so that each mRNA molecule in the cell can Count digitally. Finally, the 30dT base is synthesized, which serves as a capture region for the polyadenylation tail of the mRNA (often referred to as "oligo-dT" in the literature).
[0163] Synthesis of Uniquely Barcoded Beads
...
Embodiment 2
[0190] Example 2: Genome-wide expression profiling of thousands of individual cells using nanoliter microdroplets
[0191] Disease occurs in complex tissues made of different types of cells and (almost) never involves a single cell acting on itself: cells always interact with each other to make collective decisions, collaborate dynamics, and function together. In normal tissues, this leads to homeostasis; in disease, dysfunction of one or more interacting functions can lead to or exacerbate a pathological state.
[0192] Cells, the fundamental units of biological structure and function, vary widely in type and state. Single-cell genomics can characterize cellular identity and function, but limitations of simplicity and scale have prevented its widespread application. Applicants here describe Drop-Seq, which is a rapid method of analyzing the RNA of each cell by isolating thousands of individual cells into nanoliter-sized aqueous droplets, barcoding the RNA of each cell, and s...
Embodiment 3
[0265] Embodiment 3: the extended experiment process of embodiment 2
[0266] Bead synthesis. Bead functionalization and reverse phosphoramidite synthesis (5'-3') were performed by Chemgenes Corp. Toyopearl HW-65S resin (average particle size of 30 microns) was purchased from Tosoh Biosciences, and the surface alcohols were functionalized with PEG derivatives to create 18-carbon long flexible chain linkers. Functionalized beads were then used as solid supports for reverse phosphoramidite synthesis (5'→3') of DNA synthesis on an Expedite 8909 DNA / RNA synthesizer using a 10 μmol cycle scale and a coupling time of 3 minutes. The amides used were: N 6 -Benzoyl-3'-O-DMT-2'-deoxyadenosine-5'-cyanoethyl-N,N-diisopropyl-phosphoramidite (dA-N 6 -Bz-CEP);N 4 -Acetyl-3'-O-DMT-2'-deoxy-cytidine-5'-cyanoethyl-N,N-diisopropyl-phosphoramidite (dC-N 4 -Ac-CEP); N 2 -DMF-3′-O-DMT-2′-deoxyguanosine-5′-cyanoethyl-N, N-diisopropyl-phosphoramidite (dG-N 2 -DMF-CEP); and 3'-O-DMT-2'-deoxythy...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com