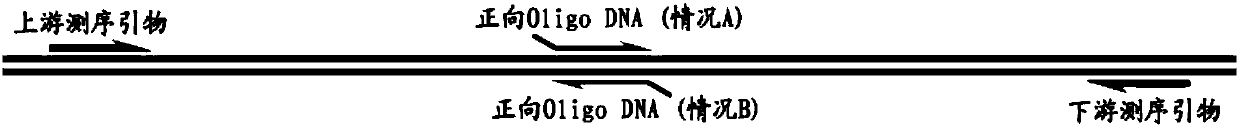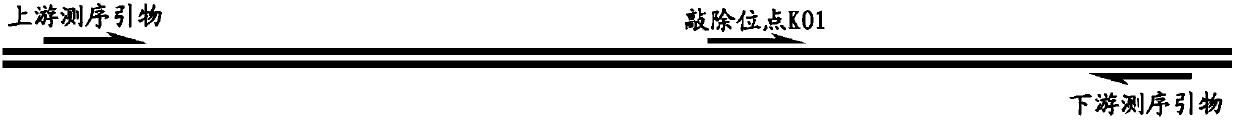Method for quickly and simply screening CRISPR/Cas gene editing positive object
A simple and convenient technology for gene editing, applied in the biological field, can solve the problems of false positives and false negatives, omission of qualified samples, increased costs, etc., to achieve the effect of reducing experimental steps and high pass rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] A quick and easy method for screening CRISPR / Cas gene editing positive objects, comprising the following steps:
[0048] Specifically, the mHnrnpa2b1 gene single point knockout screening in CHO-K1 cells
[0049] On NCBI, query the genome information of Hnrnpa2b1 (NCBI Gene ID: NM_016806.3). Design a knockout site KO1 and its corresponding Oligo DNA sequences KO1-F, KO1-R, and design upstream and downstream sequencing primers F and R ( image 3 ).
[0050] KO1-F: caccgAGCGACTGAGTCCGCGATGG
[0051] KO1-R: aaacCCATCGCGGACTCAGTCGCTc
[0052] F: GTGGGGTTAATAGCTCAGCT
[0053] R: AGAAGGAACAGGCTAAGGTG
[0054] 1) After the Oligo DNA corresponding to the knockout site KO1 was hybridized, it was connected to the BbsI site of the pSpCas9-2A-puro knockout vector to construct a plasmid pKO1.
[0055] 2) Transform Top10Competent Cell, screen positive recombinants and verify by sequencing.
[0056] 3) Large-scale extraction of plasmids and endotoxin removal for transfection;
...
Embodiment 2
[0072] A quick and easy method for screening CRISPR / Cas gene editing positive objects, comprising the following steps:
[0073] Specifically: double-site knockout screening of period1 gene in MEF cells
[0074] On NCBI, query the genome information of Period1 (NCBI Gene ID: 18626). Design two knockout sites KO1 and KO2 and their corresponding Oligo DNA sequences KO1-F, KO1-R, KO2-F, KO2-R, and design upstream and downstream sequencing primers CXF and CXR ( Figure 5 ).
[0075] KO1-F: caccgATTAGTCAGCCTCAGAGAC
[0076] KO1-R: aaacGTCTCTGAGGGCTGACTAATc
[0077] KO2-F: caccgCCCCCATCGGCCCCTTCTAG
[0078] KO2-R: aaacCTAGAAGGGGCCGATGGGGGc
[0079] CXF: GTGGGGTTAATAGCTCAGCT
[0080] CXR: AGAAGGAACAGGCTAAGGTG
[0081] 1) The Oligo DNA corresponding to the two knockout sites KO1 and KO2 were hybridized respectively, and connected to the BbsI site of the pSpCas9-2A-puro knockout vector to construct plasmids pKO-1 and pKO-2.
[0082] 2) Transform Top10Competent Cell, screen posit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com