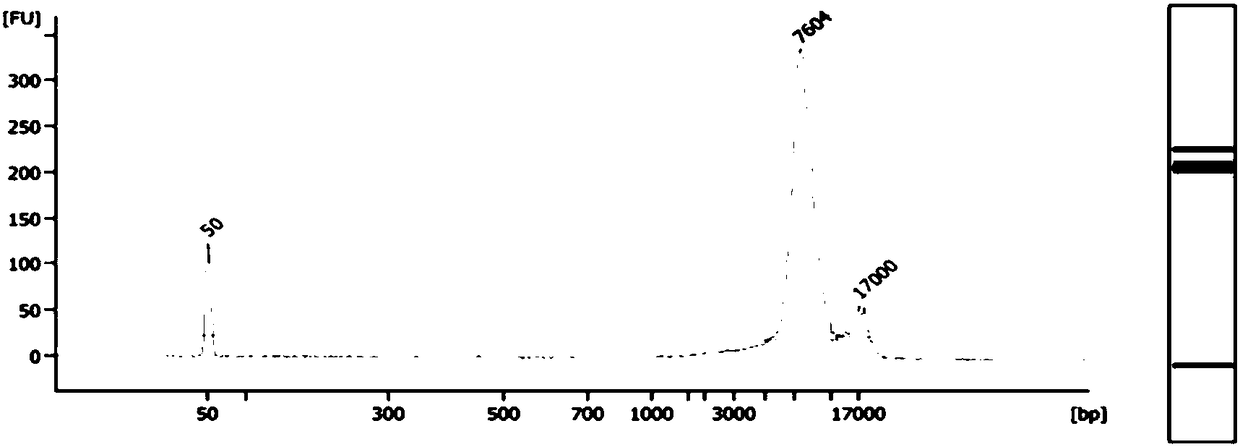
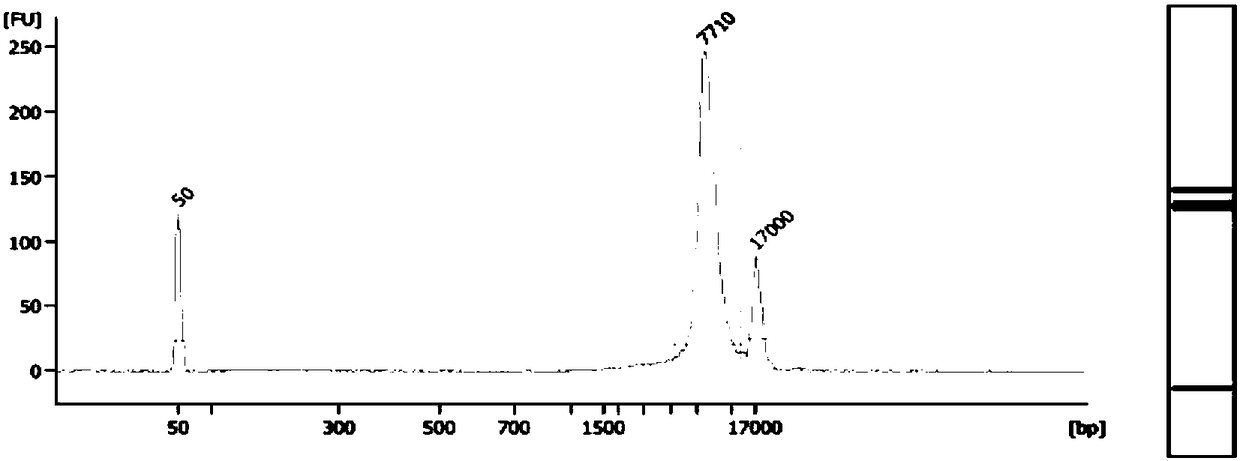
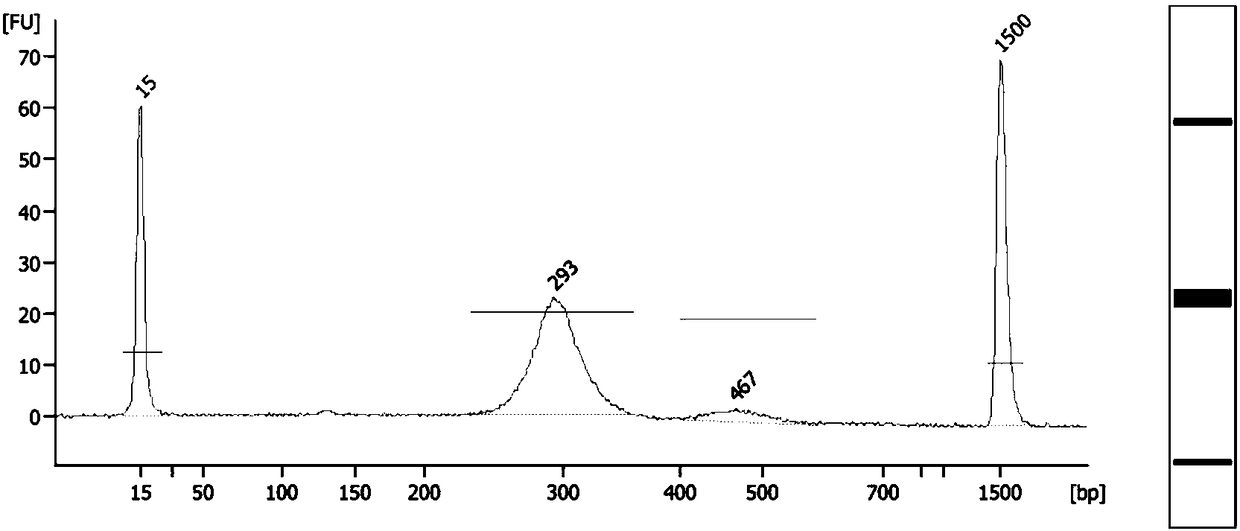
Fetus spinal muscular atrophy gene haplotype determining method and system
A technique for muscular dystrophy and haplotypes, which is applied in the fields of genomics, biochemical equipment and methods, and microbial determination/inspection, and can solve the difficulty of sampling and the inability to detect maternal mutations, which require sample size and the number of detection sites Less problems, to achieve the effect of avoiding bleeding, strong practicability, and simple samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0172] 1. Preparation of hybrid capture chip for spinal muscular atrophy gene
[0173] To determine the region where the common spinal muscular atrophy is located, the preferred region on chromosome 5 70,922,133 to 70,955,823 is the region of spinal muscular atrophy.
[0174] To remove repeated fragments, the preferred method is to remove regions with more than 200 repeated fragments, that is, if a certain fragment can be compared to more than 200 regions on the chromosome, then remove these fragments.
[0175] To obtain a chip for SMA hybrid capture, the preferred method is a liquid phase capture chip.
[0176] At the same time, the PCR method can be used to amplify and enrich the long fragments, and the PCR method can be used as a supplementary method to assist the chip in capturing the target region.
[0177] 2. Construction of the third-generation library
[0178] a. Sample preparation: Obtain blood samples from the fetal father and mother, and extract whole-genome DNA f...
Embodiment 2
[0253] Noninvasive SMA detection was performed on one patient. Both the father and mother of this case were carriers of the c.56delT mutation, and the fetus was homozygous for the c.56delT mutation.
[0254] 1. Collection and processing of father and mother samples
[0255] Using Streck blood collection tubes, 5 mL of peripheral blood was collected from the father and mother according to the standard peripheral blood collection operation. After the collection, the peripheral blood of the father and mother was separated in time according to the standard two-step centrifugation method.
[0256] 1.1 DNA extraction from plasma
[0257] The TIANamp Micro DNA Kit was used to extract the free DNA from the mother's peripheral blood plasma. The specific operation steps are as follows:
[0258] 1.1.1 Take 600 μL of the peripheral blood plasma of pregnant women into a 2 mL centrifuge tube, add 20 μL of Proteinase K solution, shake and mix well, and centrifuge briefly.
[0259] 1.1.2 A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com