Use of Smad3 protein in peripheral blood exosome as molecular marker and liver cancer detection kit
A detection kit and molecular marker technology, applied in the field of biotechnology detection, can solve problems such as imperfection, death, and difficulty in treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Extraction and identification of exosomes from human peripheral blood. Methods as below:
[0029] Human peripheral blood was collected using anticoagulant blood collection tubes, and after centrifugation (3000g×5 minutes) to remove blood cells, the supernatant was frozen in a -80°C refrigerator. When testing, remove the frozen supernatant and thaw at 4°C. Centrifuge (10000g x 60 minutes) to remove large cell debris and other substances. Take 10 μl of supernatant, add 3 μl of Exoquick reagent (purchased from System Biosciences), and let stand at 4° C. for 30 minutes. Discard the supernatant after centrifugation (3000g × 30 minutes), and discard the supernatant after centrifugation again (3000g × 5 minutes), and the obtained precipitate is exosomes.
Embodiment 2
[0031] The enzyme-linked immunosorbent assay (ELISA) was used to detect the content of exosome Smad3 in peripheral blood. The specific steps were as follows: exosomes were extracted from the peripheral blood of patients, and the protein was obtained after repeated freezing and thawing three times. The Smad3 protein content was detected according to the method steps of the Smad3 protein ELISA kit (purchased from Mlbio). The AFP data comes from the detection data generated during the clinical diagnosis and treatment of patients. The Laboratory Department of the Second Hospital of Zhejiang Medical University determined the AFP content according to the method steps of the alpha-fetoprotein assay kit (purchased from Architect).
Embodiment 3
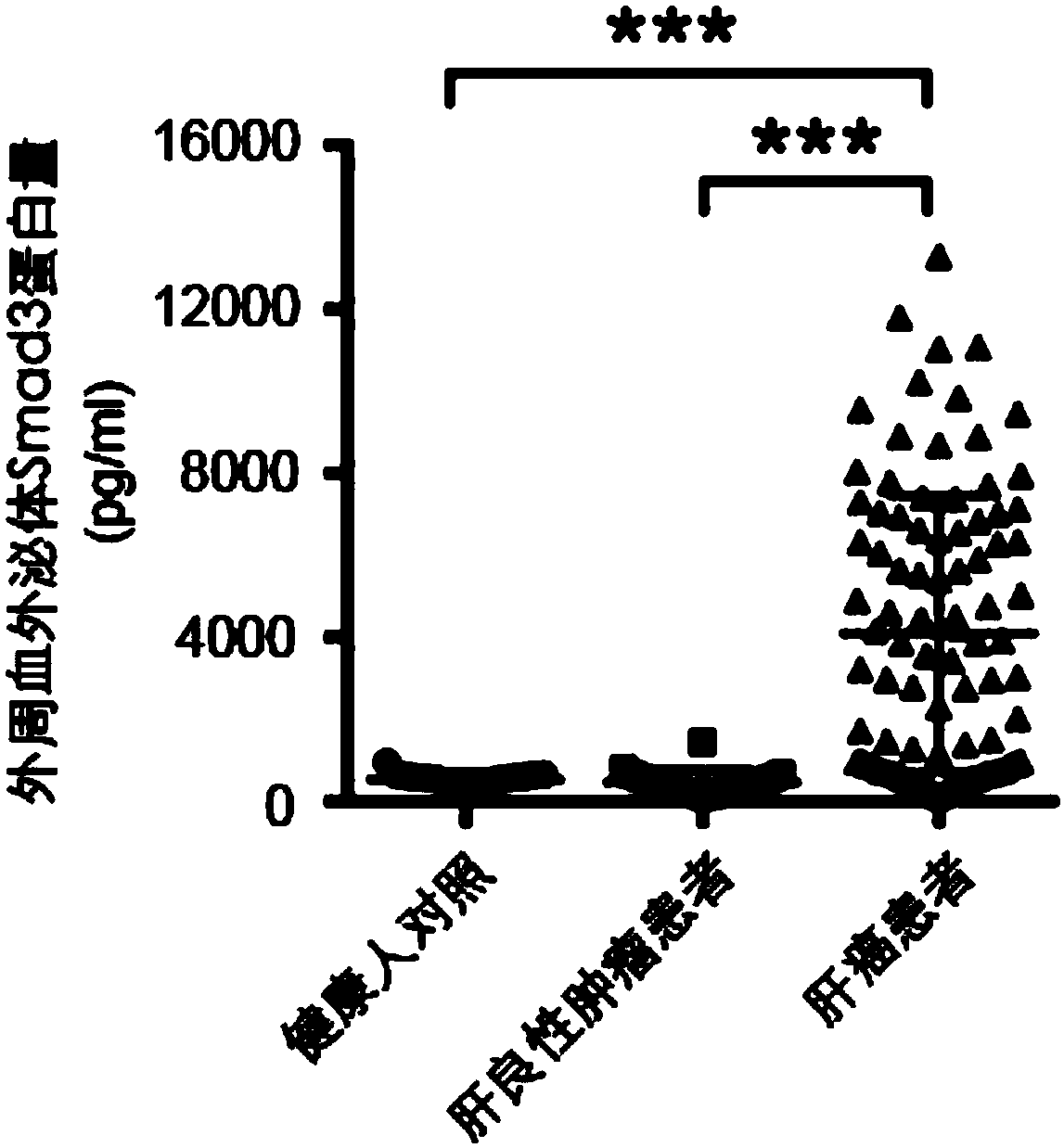
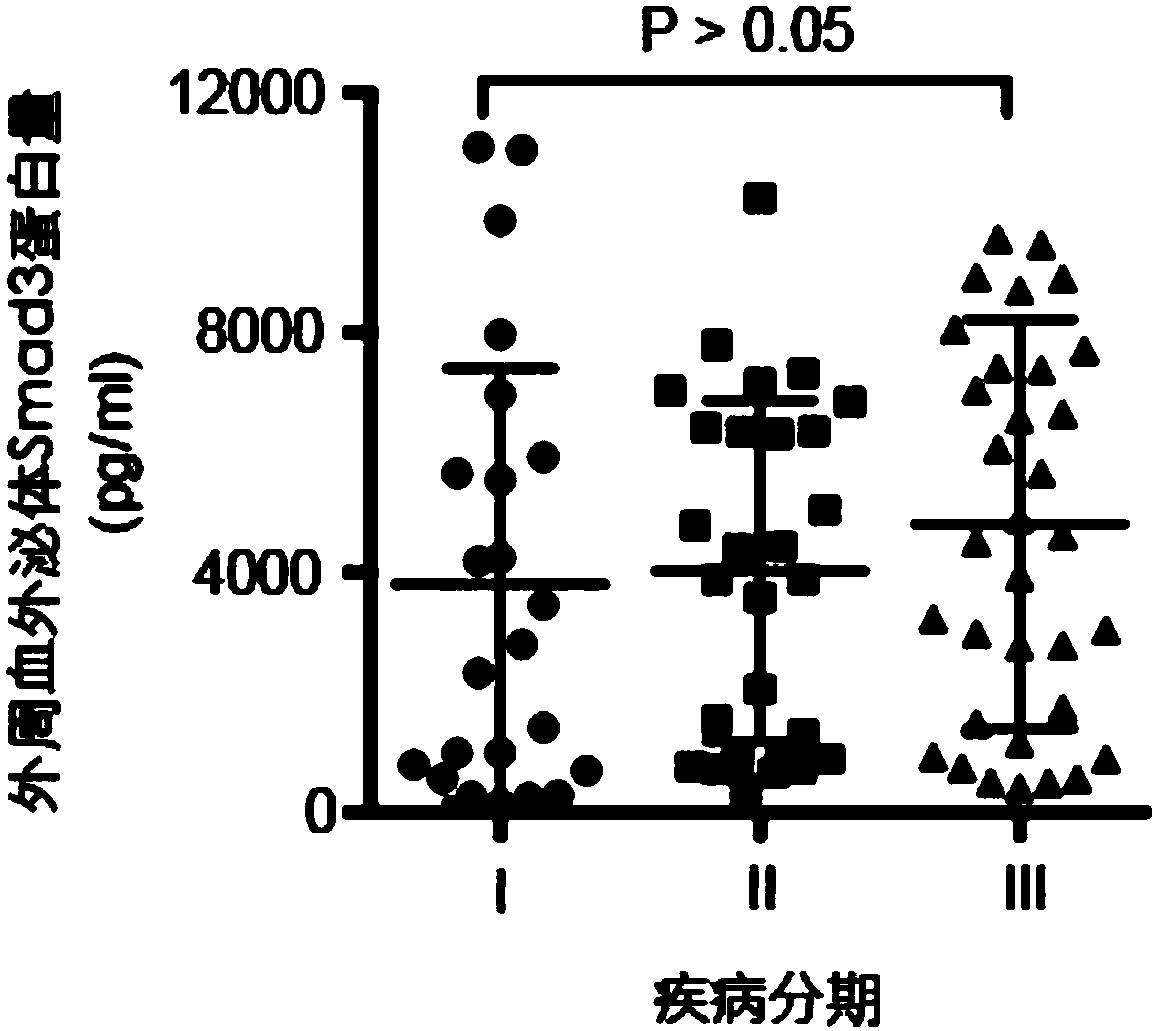
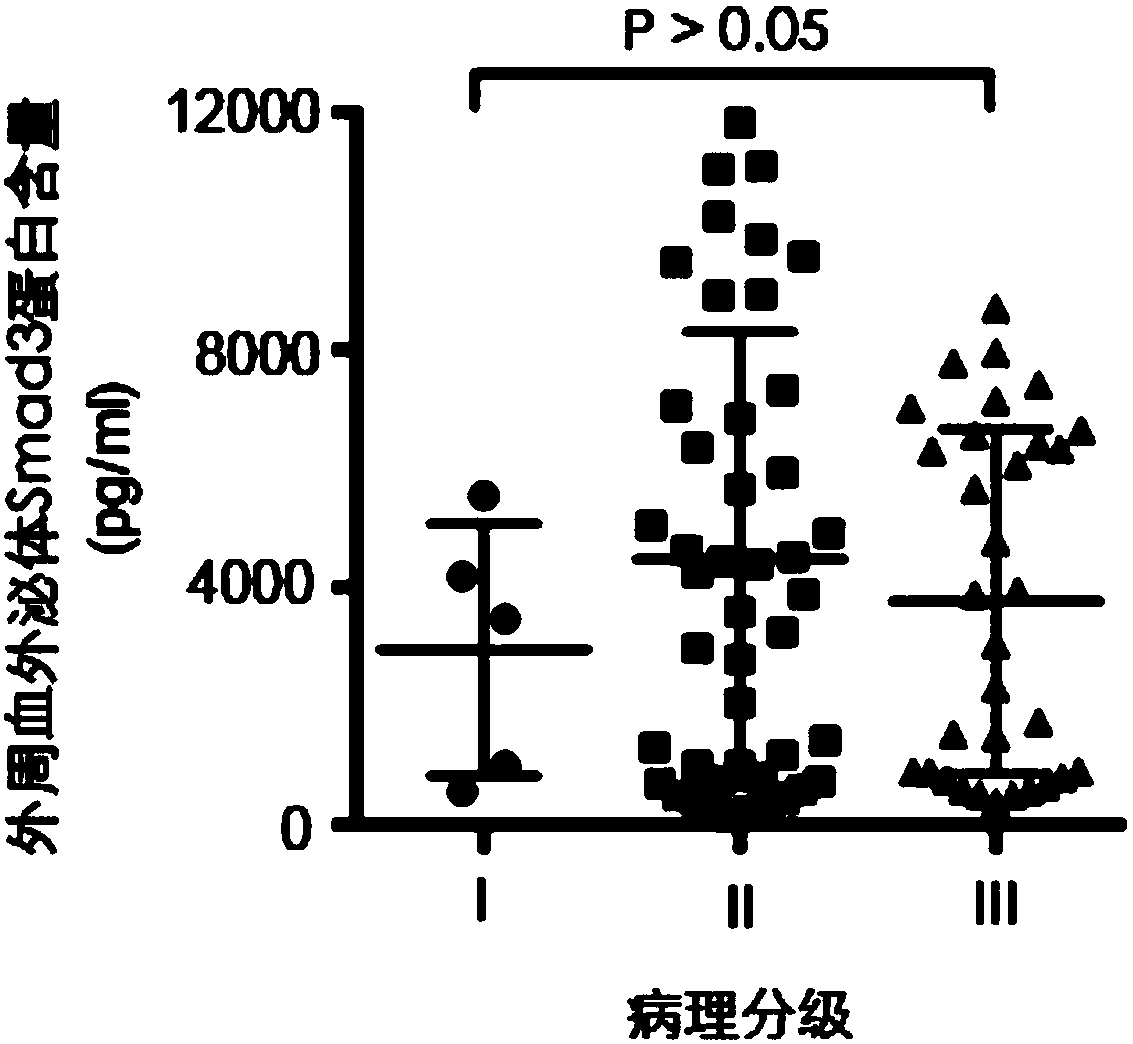
[0033] The method described in Example 2 was used to detect the Smad3 protein content in peripheral blood exosomes of patients with liver cancer and obtain corresponding AFP data, and the peripheral blood of healthy people was used as a control. The AFP data of corresponding patients and healthy people come from the detection data generated during their clinical diagnosis and treatment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com