Kit and method using mito-CRISPR/Cas9 system for knocking out DNA of abnormal mitochondria
A mito-cas9 and mito-crispr technology, applied in the field of gene editing, can solve the problems of complex operation and difficult construction of abnormal mitochondrial DNA knockout system, and achieve the effect of treating mitochondrial diseases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] 1. Selection of targeting sites
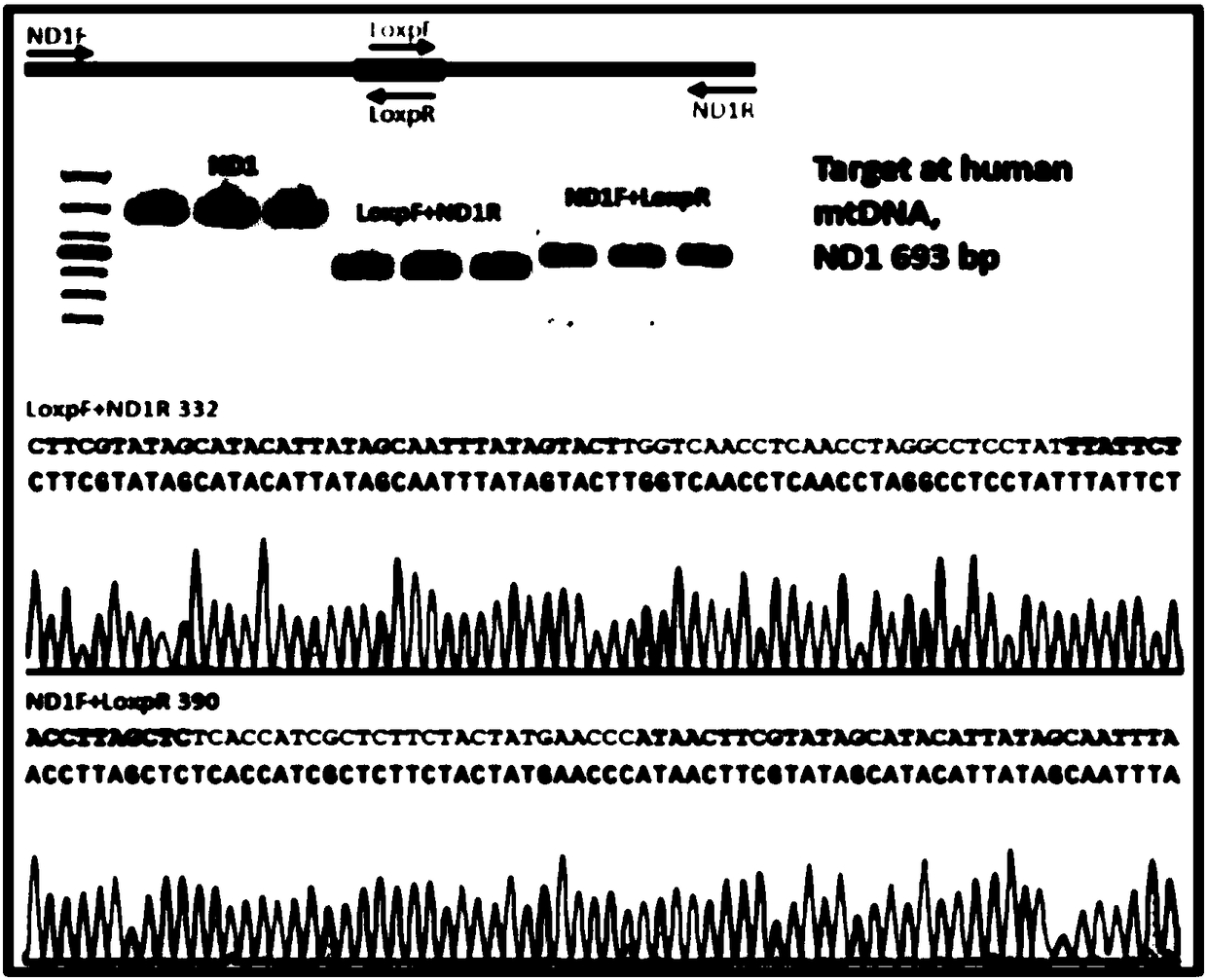
[0039] The present invention selects the ND1 gene in human mitochondria and the Dloop gene in zebrafish mitochondria. According to the editing characteristics of CRISPR / CAS9, the site is generally the 20bp sequence immediately before the NGG characteristic sequence, so the target site sequence is designed on the ZIFITTargeter (http: / / zifit.partners.org / ZiFiT / ChoiceMenu.aspx) website.
[0040] 2. Selection of relevant elements for plasmid construction
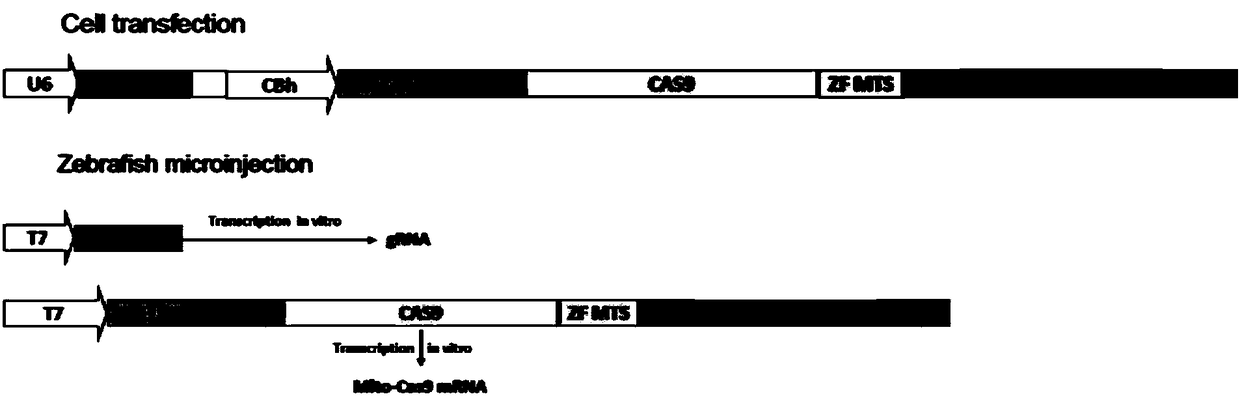
[0041] The technology for realizing mitochondrial gene modification in the present invention includes gene cloning technology, CRISPR / CAS9 gene editing technology and homologous recombination technology. In the embodiment of the present invention, the nuclear localization signal in the original vector pSpCas9(BB)-2A-eGFP is replaced by the mitochondrial localization signal by using cloning technology, so as to realize the knockout of the target gene.
[0042] In addition, the exogenous si...
Embodiment 2
[0112] Insertion of exogenous ssDNA sequences into the zebrafish mitochondrial genome by homologous recombination
[0113] Step 1: Construct the gRNA sequence of the targeting sequence
[0114] At first, the genome sequence of the Dloop gene of zebrafish was found on the website of NCBI (National Center for Biotechnology Information, USA). Find the 3'UTR region, and the locked target sequence is GCTTTGTCACATGTATGTAC.
[0115] Then, in order to construct the targeting sequence gRNA, synthesize the following primers;
[0116] Dloop-gRNA-F: TAATACGACTCACTATAGGGCTTTGTCACATGTATGTACGTTTTTAGAGCTAGAAATAGC
[0117] Dloop-gRNA-R:AGCACCGACTCGGTGCCAC
[0118] Using the plasmid with the gRNA backbone as a template, the DNA sequence of the gRNA was amplified using gRNA-F and gRNA-R at a final concentration of 10 μM as primers.
[0119] The formula is as shown in table 3:
[0120] table 3
[0121]
[0122]
[0123] PCR programming:
[0124] Step 1: 95°C for 2 minutes;
[0125] ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Outer diameter | aaaaa | aaaaa |
| The inside diameter of | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com