Method for achieving HMGCR gene knockout based on CRISPR/Cas9 technology
A gene knockout and technology technology, applied in the field of genetic engineering, can solve problems such as HMGCR gene knockout that have not yet been seen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
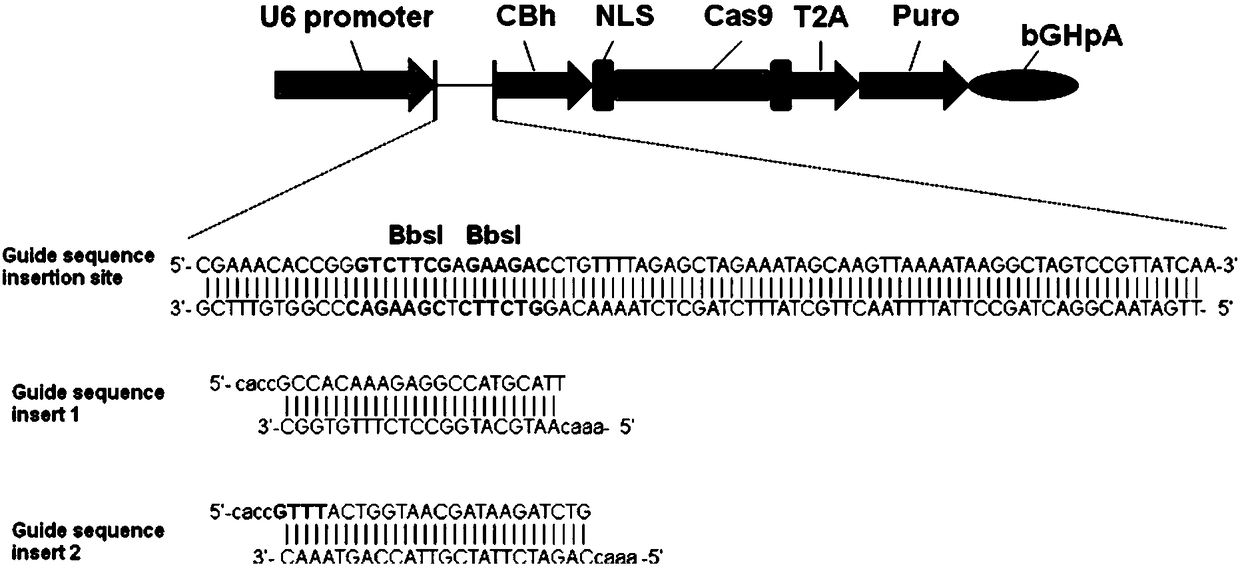
[0029] Embodiment 1: vector construction
[0030] (1) HMGCR target optimization design
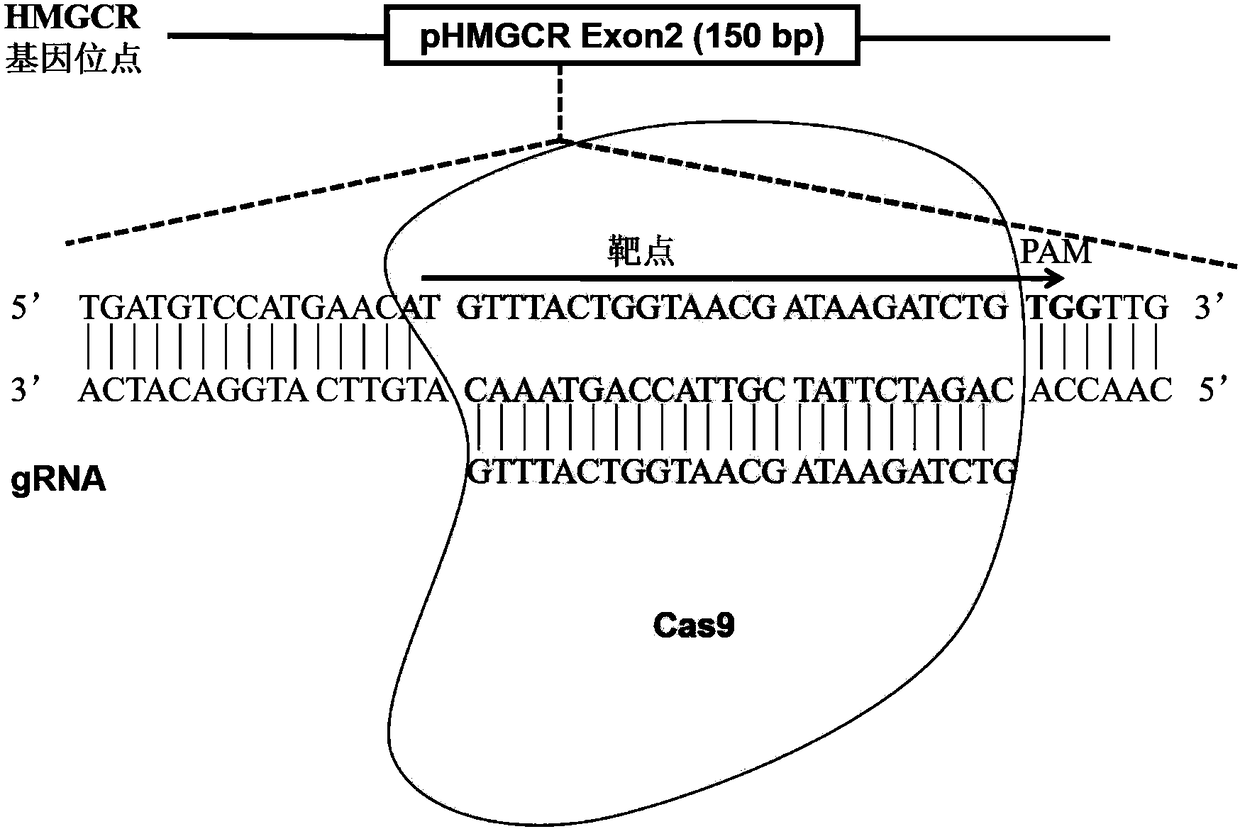
[0031]For the HMGCR gene (gene name HMGCR, gene ID number: 100144446, see https: / / www.ncbi.nlm.nih.gov / gene / ?term=DQ432054.1 for gene details), retrieve and download HMGCR from the Genebank website Partial genome sequence (SEQ ID NO: 1): TTCTGAAgCTACAATGTTGTCAAGACTTCTTCCGAATGCATGGCCTCTTTGTGGCCTCCCATCCCTGGGAAGTCATAGTGGGGACAGTGACACTGACCATCTGTATGATGTCCATGAACATGTTTACTGGTAACGATAAGATCTGTGGTTG.
[0032] Using the online software Feng Zhang lab's Target Finder( http: / / crispr.mit.edu / ) to design gRNA, input the 150bp of the above HMGCR genome sequence, set and retrieve several gRNA sequences, and analyze the position of the gRNA on the gene sequence and the off-target (off-target) information of the gRNA, see figure 1 , respectively select the optimal upstream target sequence, as shown in SEQ ID NO: 3; the downstream 1 target sequence, as shown in SEQ ID NO: 2, specifically as follows:
[0033...
Embodiment 2
[0083] Example 2: Test knockout efficiency and construct HMGCR knockout PK15 cell line
[0084] (1) Plasmid amplification
[0085] 1) Take out 100ul competent cells from the ultra-low temperature refrigerator at -80°C, place them on ice, and gently suspend the cells evenly after thawing completely;
[0086]2) Add 1 μl of plasmid (PX459-HMGCR-gRNA1, PX459-HMGCR-gRNA2) and mix gently, and place on ice for 30 minutes;
[0087] 3) Heat shock in a water bath at 42°C for 60 seconds, and place on ice for 2 minutes;
[0088] 4) Add 500 μl of SOC medium (containing MgCL2), culture at 37°C, 225rpm for 1 hour to recover;
[0089] 5) Mix the bacterial solution with a pipette tip, take 100 μl (up to 200 μl), and spread the bacteria on the ampicillin plate;
[0090] 6) The plate was placed upright at 37° C. for 10 minutes to absorb excess liquid, and then cultured upside down overnight (about 12 hours).
[0091] 7) Pick a single colony from the ampicillin culture plate, infuse it into a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com