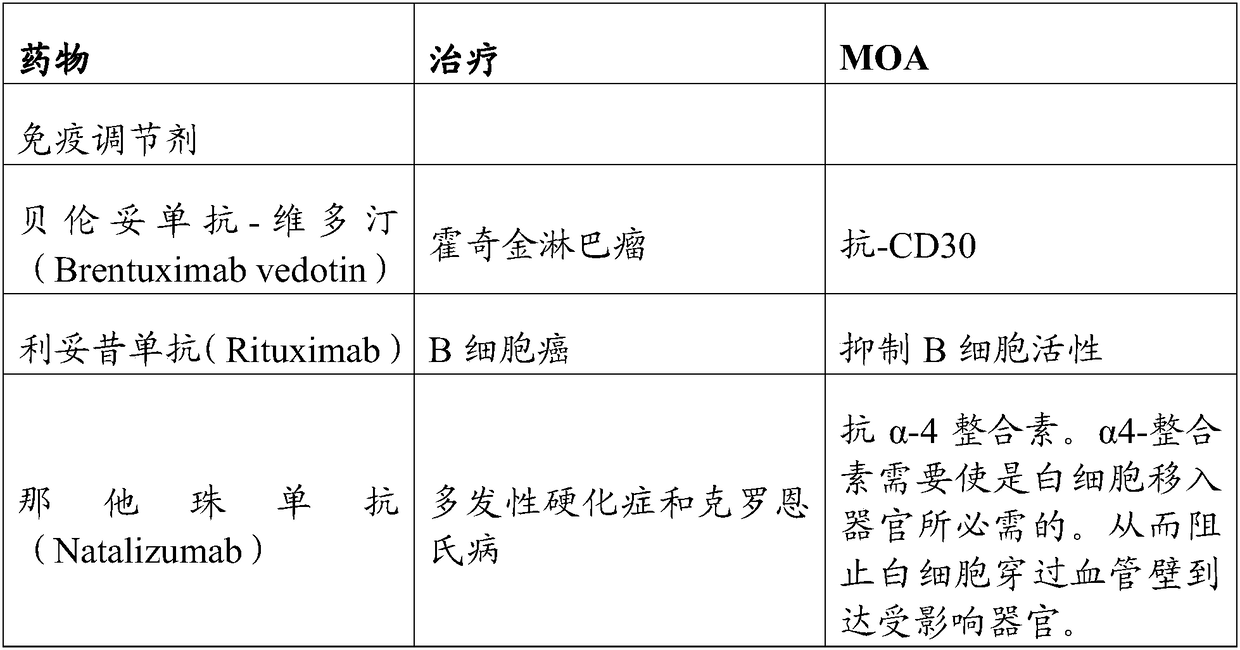
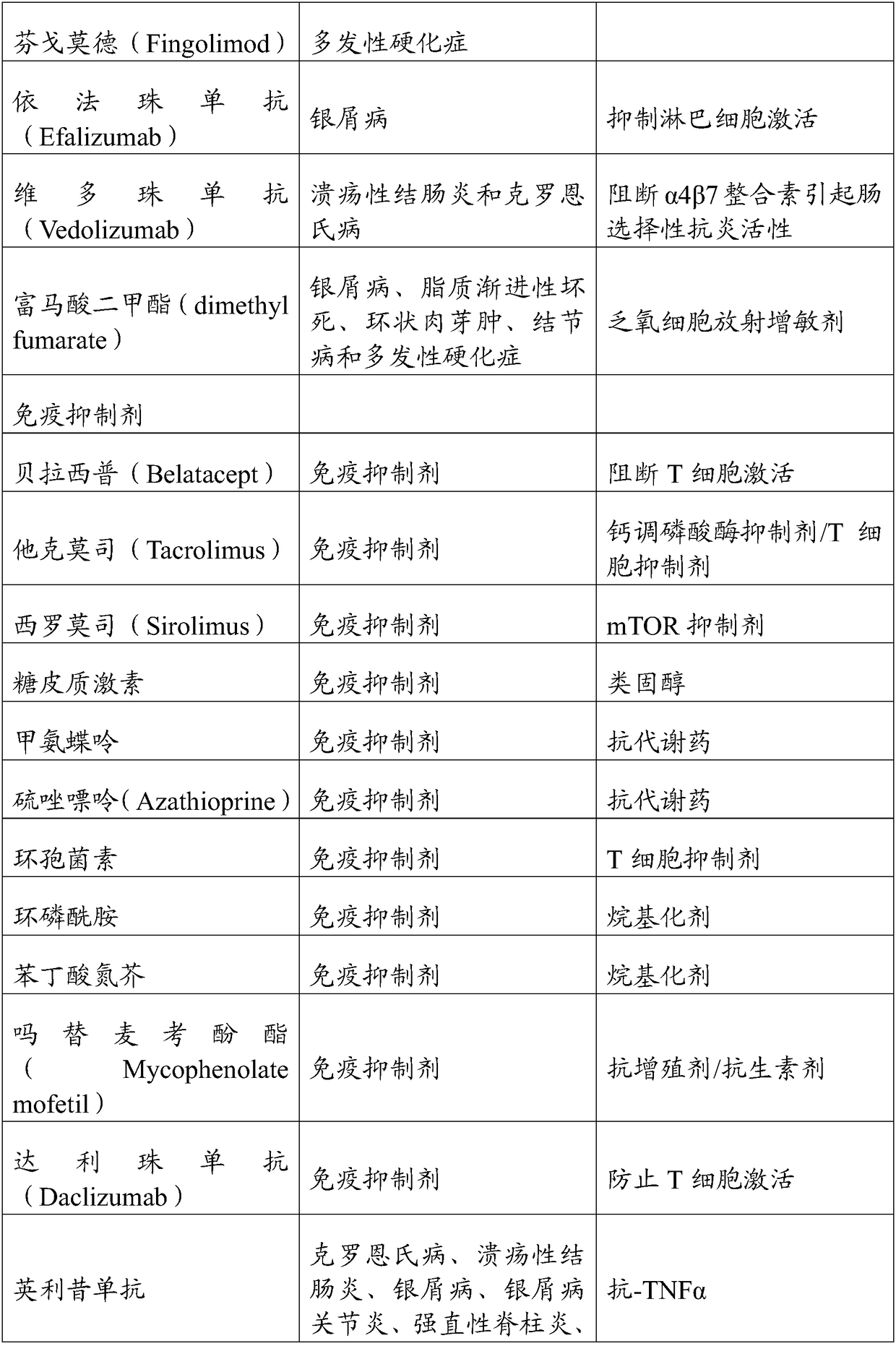
Gene editing methods and compositions for eliminating risk of jc virus activation and pml (progressive multifocal leukoencephalopathy) during immunosuppresive therapy
A technology of immunosuppression and gene editing, applied in the direction of immunoglobulin, chemical instruments and methods, biochemical equipment and methods, etc., can solve problems such as deletion, insertion and excision of DNA fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0039] The gRNAs in Example 1 are those disclosed by Wollebo et al. (2015), but it should be understood that the present invention is not limited to those gRNAs. The gRNA includes a gRNA spacer sequence complementary to the JCV T-antigen sequence in the TM1, TM2 or TM3 region. The target sequence can extend from about 20 to 40 or more nucleotides (nt) in length. It should be understood that in different JCV strains or mutant variants, sequences homologous to TM1, TM2 and TM3 can be easily identified by well-known sequencing and genomic techniques.
[0040] An exemplary target sequence in TM1 includes SEQ ID NO: 1 or its complementary sequence SEQ ID NO: 2 on the antiparallel chain. The target sequence can include the PAM sequence in each chain (in figure 1 (Shown in bold and lowercase letters), so that the target sequence may include SEQ ID NO: 3 or its complementary sequence SEQ ID NO: 4 on the antiparallel strand. Therefore, the gRNA complementary to TM1 designated as gRNAm1 ...
example 2
[0109] Example 2: CRISPR / Cpf1 composition and method for elimination of JCV as a co-therapeutic treatment with natalizumab.
[0110] Based on the 3'neighbor of the 5'TTN sequence in the JCV T-Ag genome, the hypothetical target sequence of Cpf1 is disclosed in Table 2 (as the target sequence cm1-cm236). The gene editing composition of the present invention includes at least one gRNA complementary to one of the listed target sequences. The gRNA of the present invention may or may not include a sequence complementary to the PAM sequence of the target sequence, which is listed in brackets at the 5'end of each target sequence in Table 2. The gRNA may be complementary to truncated variants of the listed sequence, for example, a sequence that has been truncated by 1, 2, 3, or more nucleotides at the 3'end. The gRNA may be less than 100% complementary to the target sequence listed in Table 2. For example, gRNA can be 95% complementary to the listed target sequence. The gRNA sequence m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com