Genome editing in bacillus host cells
A host cell and bacillus technology, applied in genetic engineering, virus/bacteriophage, recombinant DNA technology, etc., can solve problems such as the successful application of bacillus host cells that have not yet been reported
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
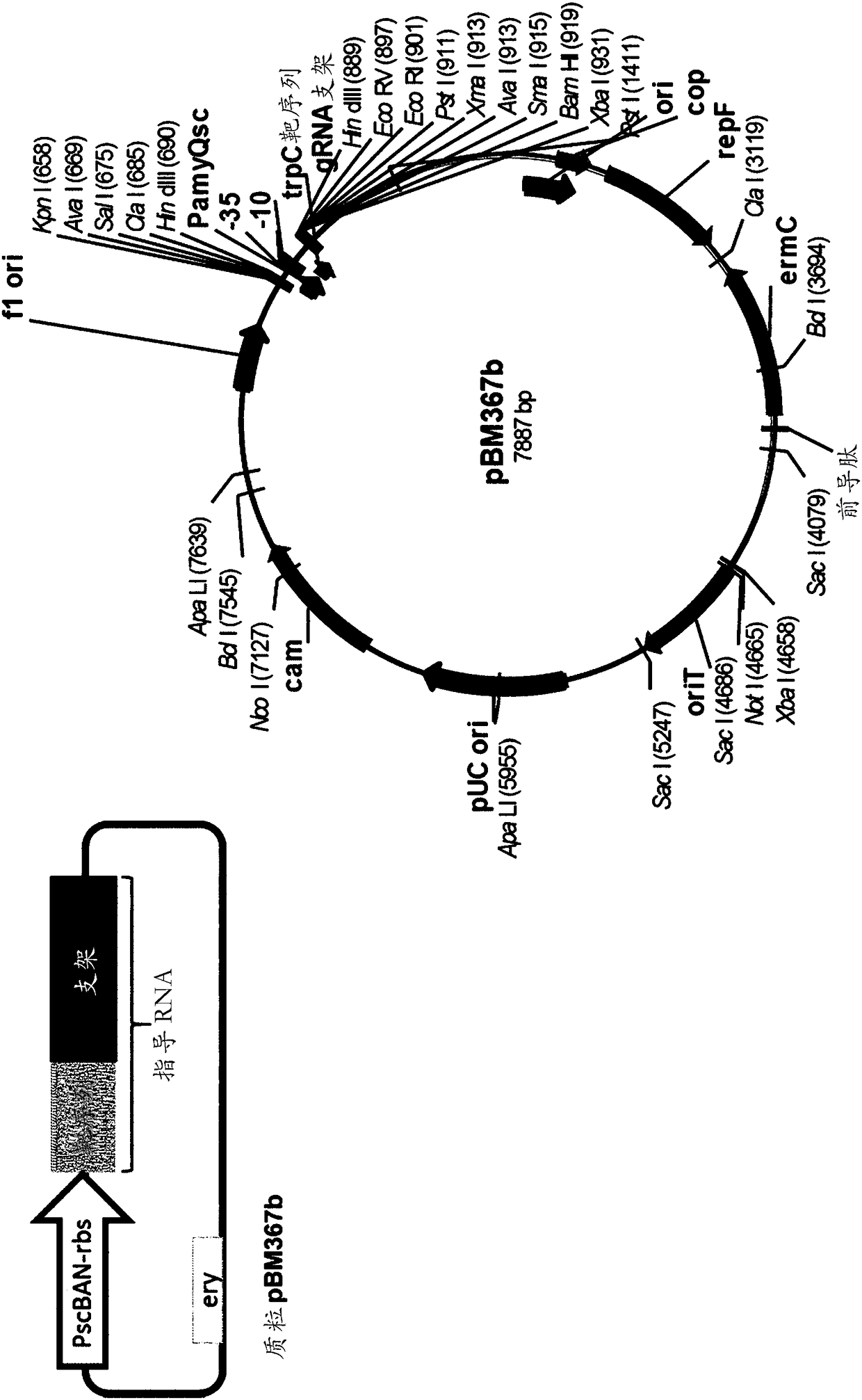
[0148] Example 1. Construction of plasmid pBM353
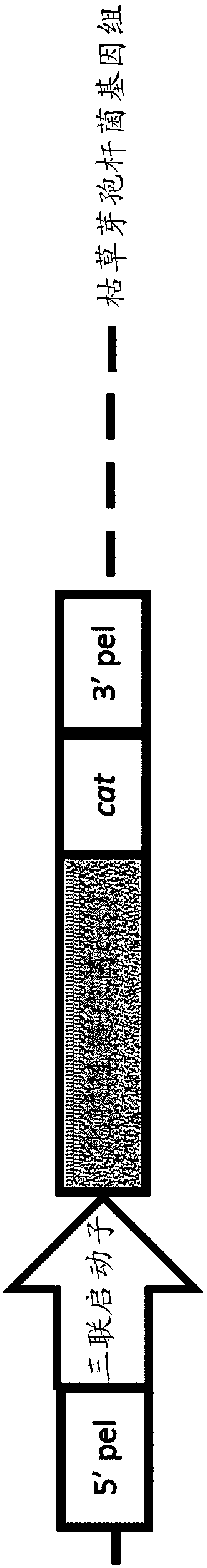
[0149] Plasmid pBM353 was designed to simultaneously disrupt parts of the ykoV (ligD) and ykoU (ku) genes in B. subtilis. Since these two genes are in the same operon, this plasmid was designed to delete the C-terminus from ykoV (retained amino acids 1-174; total protein is 311 amino acids) and remove the ku gene (612 amino acids full-length protein). remove the first 28 amino acids.
[0150]According to the aforementioned method (Pitcher, D.G., N.A.Saunders and R.J.Owen, Rapid extraction of bacterial genomic DNA with guanidium thiocyanate [quick extraction of bacterial genomic DNA with guanidinium thiocyanate]. Letters in Applied Microbiology [Application Microbiology Newsletter], 1989.8 (4 ): pp. 151-156) Isolation of genomic DNA from Bacillus subtilis 168.Δ.4. A 555 bp fragment of the Bacillus subtilis 168.Δ.4 chromosome was amplified by PCR using primers 1213241 and 1213243 shown below.
[0151] Primer 1213241 (SEQ ID N...
example 2
[0161] Example 2. Construction of plasmid pBM354
[0162] A temperature-sensitive Bacillus / E. coli shuttle vector based on pShV002, which does not contain a BsaI restriction site, was generated using site-directed mutagenesis. Use the following primers for the PCR reaction:
[0163] Primer 1213365 (SEQ ID NO:5): 5'-gctgaataaaagatacgaagacctctcttgtatct
[0164] Primer 1213366 (SEQ ID NO:6): 5'-agatacaagagagggtcttcgtatcttttattcagc
[0165] Plasmids were amplified by PCR using the Agilent Technologies Quickchange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). The PCR amplification reaction mixture contained 1 μl of 21 ng / μl pShV002, 1 μl of sense primer (50 pmol / μl), 1 μl of antisense primer (50 pmol / μl), 5 μl of 10X reaction buffer, 1 μl of dNTP mix (each 10 mM ), 3 μl of Quick solution, 37 μl of water and 1 μl (2.5 U / μl) of PfuUltra HF DNA polymerase. Fragments were amplified using an Eppendorf Mastercycler thermal cycler with the following sett...
example 3
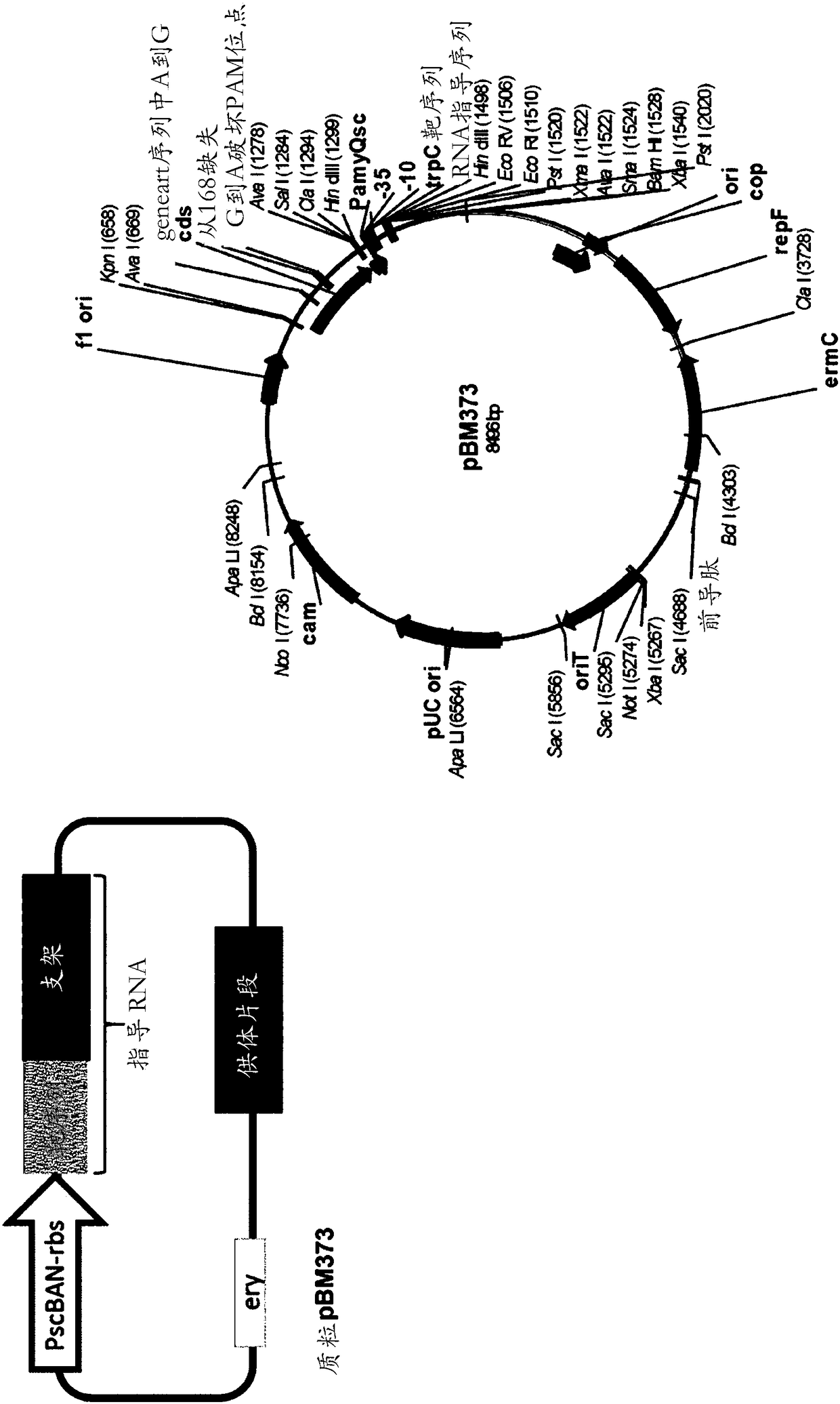
[0166] Example 3. Construction of plasmid pBM363b
[0167] A synthetic DNA fragment containing the S. pyogenes cas9 gene was obtained from GeneArt (Thermo Fischer Scientific, Grand Island, NY); the DNA sequence is provided in SEQ ID NO: encoding SEQ ID NO:8 7 in. This fragment was cloned into the temperature sensitive Bacillus / E. coli shuttle vector pBM354 as follows.
[0168] The following primers were used to amplify the cas9 gene:
[0169] Primer 1213801 (SEQ ID NO:9): 5'-gaattgggtaccgggccccccctcgagtcgacatgccggtactgccg
[0170] Primer 1213802 (SEQ ID NO: 10): 5'-cgatatcaagcttatcgataccgtcgacgtgactggcgatgctgtcgg
[0171] The corresponding DNA fragments were amplified by PCR using the Expand High Fidelity PLUS PCR system (Roche Diagnostics, Mannheim, Germany). The PCR amplification reaction mixture contained 1 μl of 0.05 μg / μl synthetic DNA, 1 μl of sense primer (50 pmol / μl), 1 μl of antisense primer (50 pmol / μl), 10 μl of DNA with 15 mM MgCl 25X PCR buffer, 1 μl of dNTP ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com