Mini-ILL RNASES, methods for changing specificity of RNA sequence cleavage by Mini-ILL RNASES, and uses thereof
A specificity and sequence technology, applied in Mini-III RNase, changing the specificity of Mini-III RNase cleaving RNA sequence and its application field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
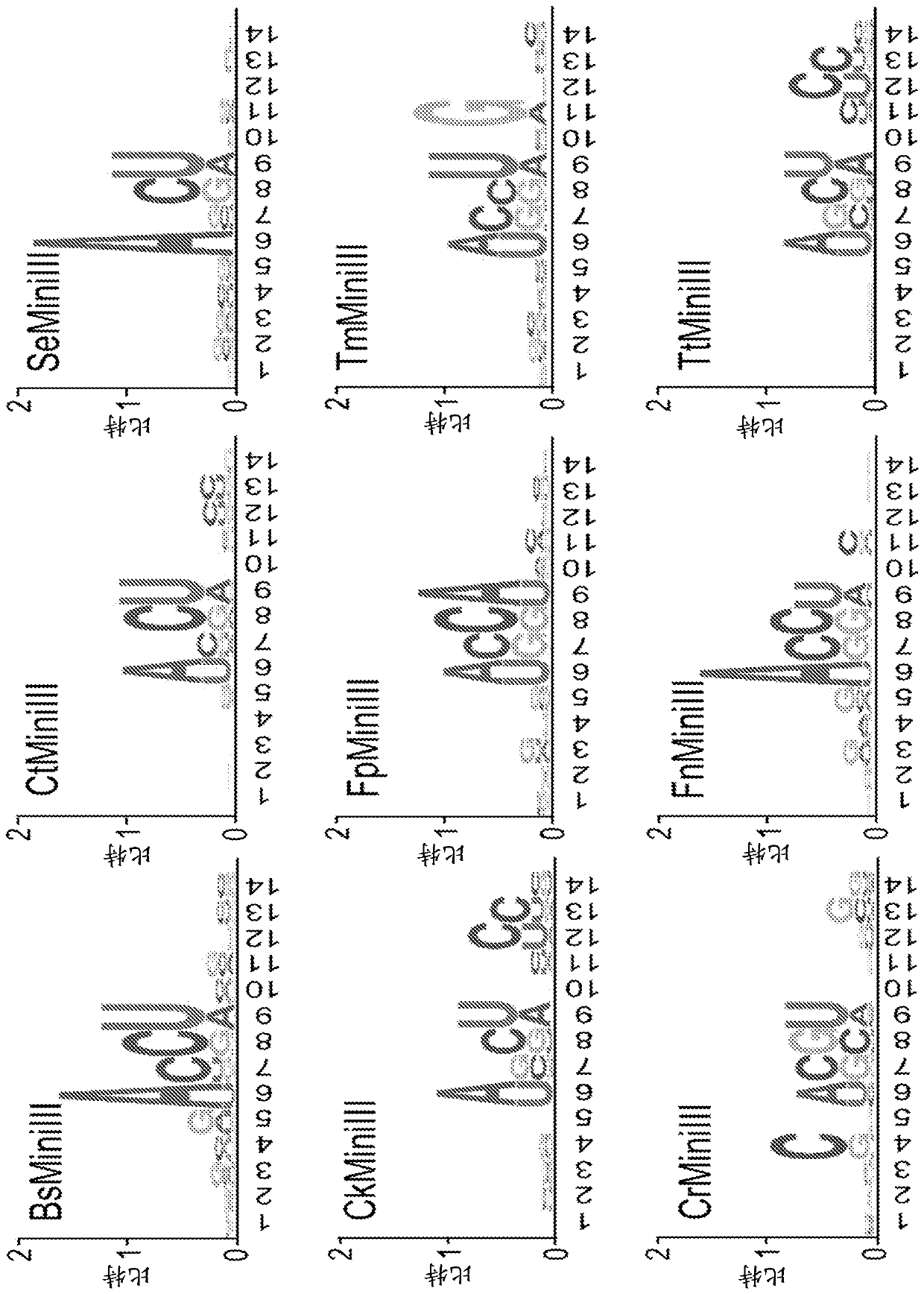
[0147] Cloning of sequences encoding specific Mini-III RNases
[0148] Microorganisms were purchased in lyophilized form from DSMZ (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures) (Table 1). After suspending the lyophilized product in 500 µl of TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0), the suspension was extracted with phenol (saturated with 100 mM Tris-HCl, pH 8.5). The aqueous phase was re-extracted with a phenol:chloroform (1 / 1 v / v) mixture, followed by nucleic acid precipitation by adding 50 μl of 3M sodium acetate pH 5.2 and 1 mL of ethanol. The precipitated nucleic acid was centrifuged (12000g, 10 min, 4°C) and the liquid was removed. The precipitate was washed with 1 mL of 70% ethanol and dried. The obtained DNA was suspended in 20 µl of TE buffer.
[0149] Table 1. Origin of cloned sequences encoding Mini-III RNases
[0150]
[0151] Table 2. Primer sequences used to amplify genomic DNA encoding specific Mini-III RNases...
Embodiment 2
[0157] Protein was expressed and purified from a recombinant plasmid encoding wild-type Mini-III RNase.
[0158] Transform Escherichia coli strain BL21 (DE3) (F–ompT gal dcm lon hsdSB(rB- mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1sam7 ) with the recombinant plasmid carrying the Mini-III nuclease gene obtained in Example 1 nin5]). Transformation was carried out according to Example 1. Transformants were selected on LB solid medium supplemented with 50 μg / mL kanamycin and 1% glucose. Inoculate selected transformant colonies in LB liquid medium and incubate with shaking at 37°C for 5 hours. Then, inoculate in 500 mL liquid ZY (Studier 2005) supplemented with kanamycin to a concentration of 100 μg / mL 25 mL of culture grown in LB medium and incubated with shaking at 37 °C for 24 h. The culture was centrifuged at 5000 g for 10 min at 4 °C and suspended in STE buffer (0.1 M NaCl, 10 mM Tris-HCl pH 8.0 , 1mM EDTA pH 8.0), and centrifuged again. The pellet was suspended in 20mL lysis so...
Embodiment 3
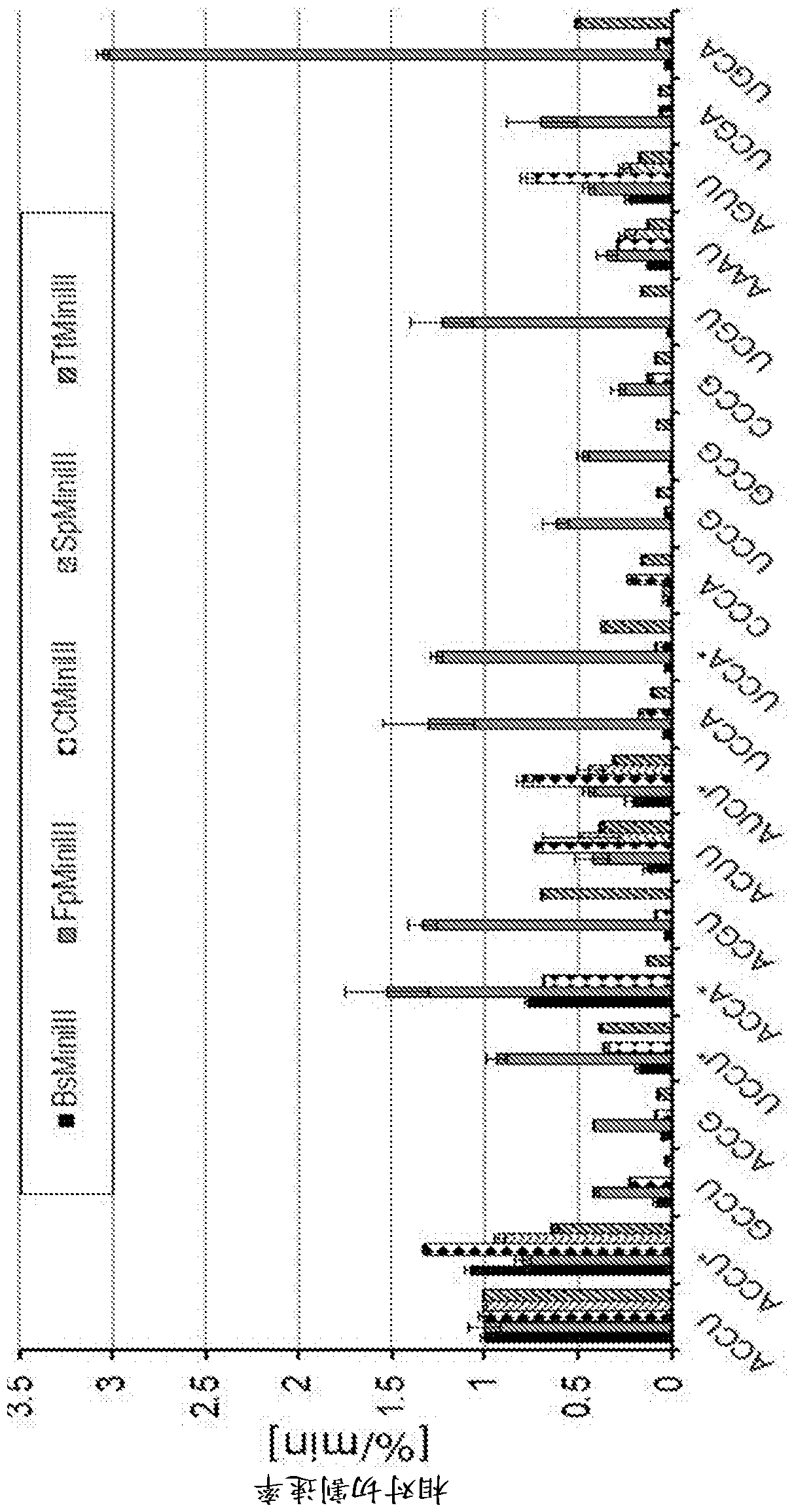
[0162] Determining optimal reaction conditions for in vitro cleavage of dsRNA substrates with purified enzymes
[0163] To determine the optimal conditions for the reaction buffer, limited cleavage of the Φ6 phage genome was performed in the buffers listed in Table 3.
[0164] table 3
[0165]
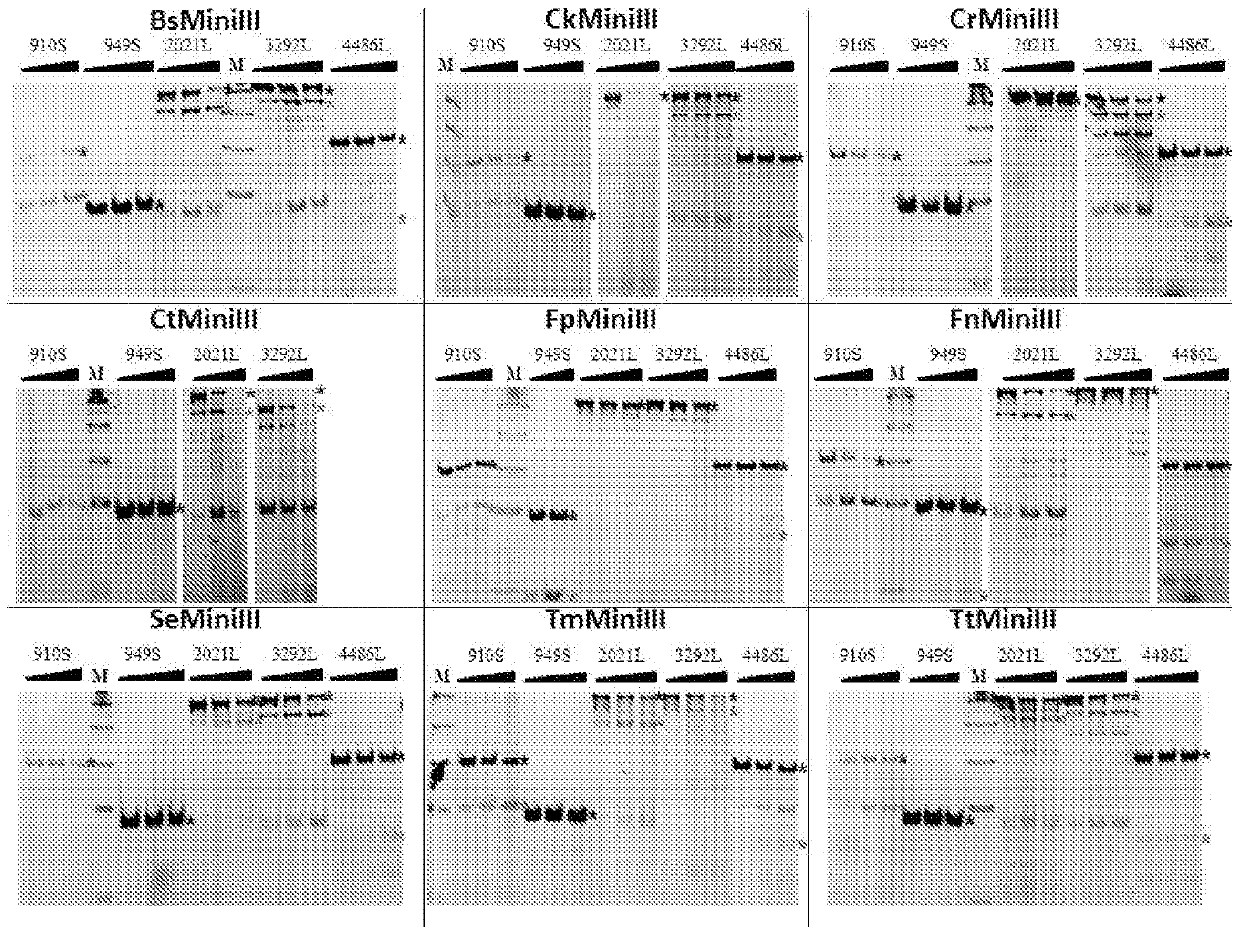
[0166] In this experiment, 1.5 μg of dsRNA was used, as well as 3.3 μg of BsMiniIII obtained in Example 2, 80 ng of CkMiniIII, 23.5 μg of CrMiniIII, 5 μg of CtMiniIII, 0.8 μg of FnMiniIII, 1.1 μg of FpMiniIII, 2.1 μg of SeMiniIII, 0.185 μg of TmMiniIII, 11.5 ng of TtMiniIII. Aliquots corresponding to 0.5 μg dsRNA were collected after 5, 10 and 15 minutes of reaction, except for TtMiniIII and SeMiniIII, which had reaction times of 2, 4 and 6 minutes, and 20, 40 and 60 minutes, respectively.
[0167] The cleavage products were separated by electrophoresis on a 1.5% agarose gel supplemented with ethidium bromide at a final concentration of 0.5 μg / mL. The buffer that produced the mos...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com