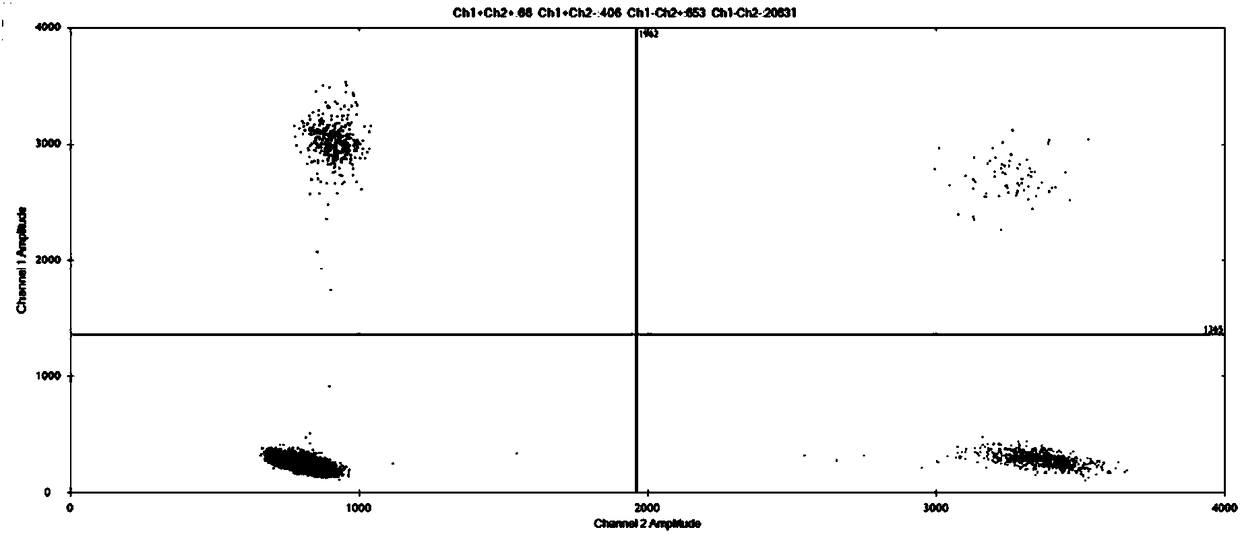
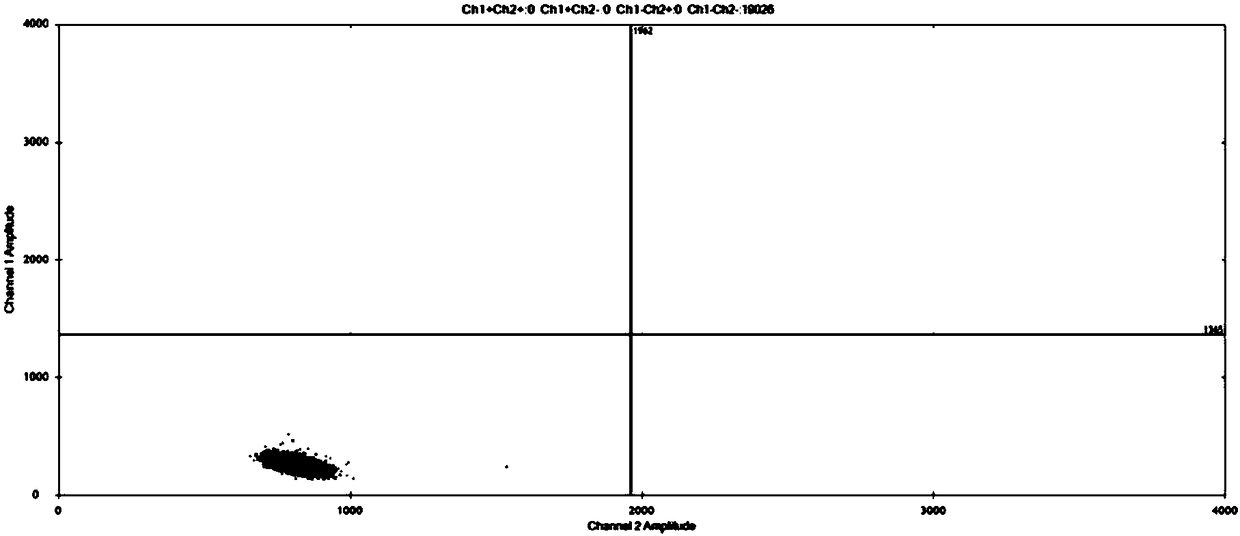
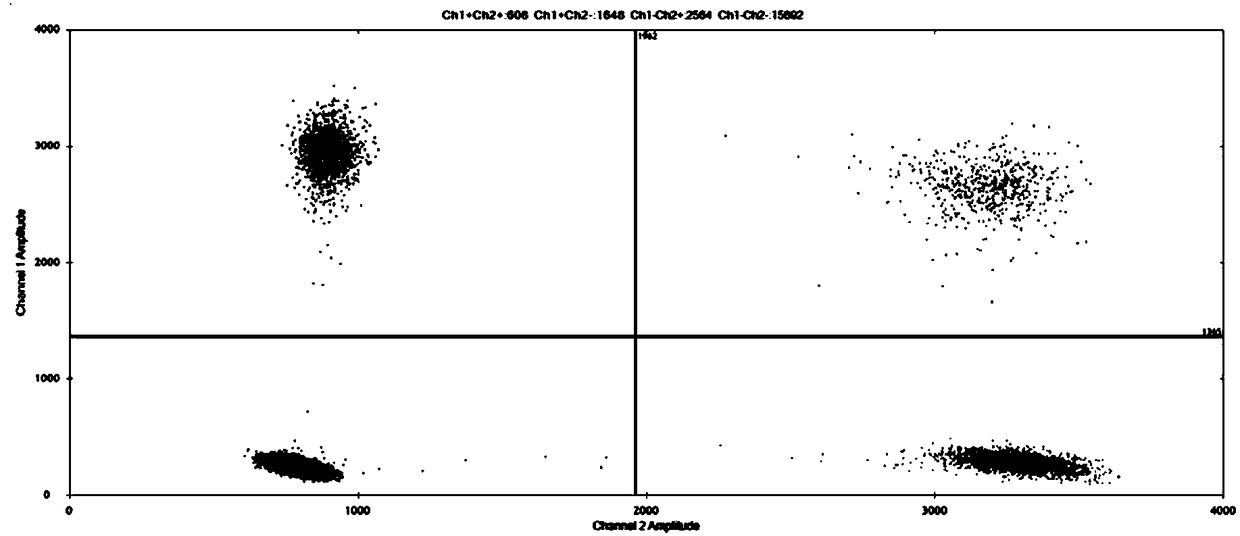
Method for improving sensitivity of digital PCR and kit thereof
A kit and digital technology, applied in the method and its kit to improve the sensitivity of digital PCR, can solve the problems of limiting the value of ddPCR and difficult detection, so as to improve the success rate, enhance the value, and improve the low detection rate of trace amount. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0107] Example 1 Preamplification Kit
[0108] The composition of the pre-amplification kit is shown in Table 9:
[0109] Table 9
[0110]
[0111]
[0112] The specific preparation method of the kit is:
[0113] ①Materials: primers and process water
[0114] ② Primer dilution:
[0115] Centrifuge all the primers instantaneously, add process water in proportion to make it dissolve, the concentration is 100 μmol, mix thoroughly, and set aside.
[0116] ③ Preparation:
[0117] Strictly follow the concentration and dosage in the above table, accurately draw the corresponding amount of each primer, prepare the pre-amplification reaction solution, 10 μL for each test, and use process water for the insufficient part. Mix well. Label the date of preparation, product name, volume, and batch number (all the raw materials used above must pass the inspection).
[0118] ④Storage conditions: The prepared solution is stored at 4°C and protected from light, and sent to quality i...
Embodiment 2
[0119] The usage method of embodiment 2 kit
[0120] 1. System preparation
[0121] Take out the corresponding reaction solution, melt and mix it at room temperature, centrifuge at 2000rpm for 10s, and prepare the PCR premix solution for each test as follows: 1.5 μL pre-amplification reaction solution + 7.5 μL S-MIX, and the PCR premix prepared above The mixed solution was dispensed into each PCR tube in an amount of 9 μL per tube.
[0122] Take 6 μL of cfDNA and the control substance in the kit and add them to the EP tubes containing the master mix.
[0123] 2. Pre-amplification procedure:
[0124] 42°C, 5min; 94°C, 5min; (94°C, 15sec; 60°C, 25sec; 72°C, 40sec) 8 cycles; 98°C, 10min; 4°C, 5min, the reaction system was set to 20μL.
[0125] 3. Take 1 μL of the pre-amplified product as a template for ddPCR reaction.
Embodiment 3
[0127] A kind of s-dd PCR kit that detects human EGFR gene mutation, its composition is as shown in table 10:
[0128] Table 10:
[0129]
[0130] The packaging and content of each component of the S-ddPCR kit are shown in Table 11:
[0131] Table 11:
[0132]
[0133]
[0134] The using method of kit of the present invention is:
[0135] 1. ddPCR detection
[0136] 1.1 Pre-amplification:
[0137] 1.1.1 Take out the pre-amplification reaction solution, thaw at room temperature and mix well, centrifuge at 2000rpm for 10s, prepare the PCR master mix for each test as follows: 1.5μL pre-amplification reaction solution + 7.5μL S-MIX, mix the above A good PCR master mix was dispensed into each PCR tube in an amount of 9 μL per tube.
[0138] Take 6 μL of cfDNA and the control substance in the kit and add them to the EP tubes containing the master mix.
[0139] 1.1.2. Pre-amplification procedure:
[0140] 42°C, 5min; 94°C, 5min; (94°C, 15sec; 60°C, 25sec; 72°C, 40se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com