Primer pair for detecting sweet potato stem rot pathogen as well as detection method thereof
A primer pair, sweet potato technology, applied in the biological field, can solve the problems of inapplicability of specific detection, difficulty in distinguishing different species of Dickeya, and difficulty in detection of sweet potato stem rot fungus, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
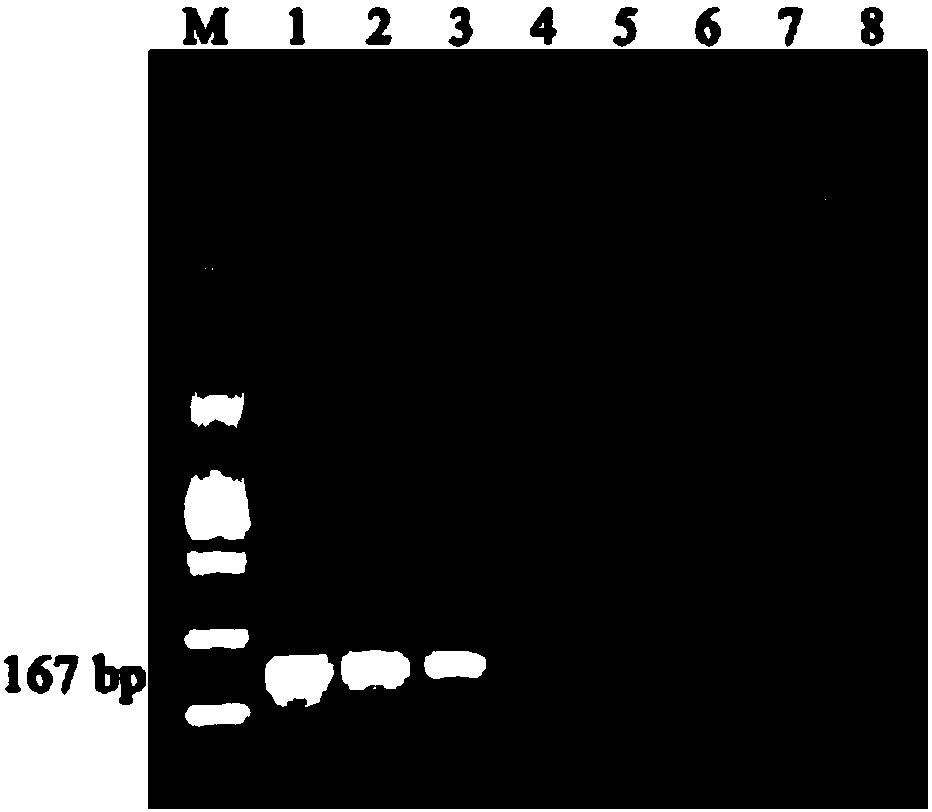
[0035] Specific gene sequence screening and primer design
[0036] 1) Specific gene screening
[0037]According to the genome sequence of P. spp. in the NCBI genome database, its specific genes were found out by pan-genome analysis, and the specificity of the found genes was verified by local Blast. The specific gene sequence is shown in SEQ ID No.1.
[0038] 2) Primer design
[0039] For the specific gene sequence, the primers are designed by Primer-BLAST, an online design specific oligonucleotide primer tool. The upstream primer is shown in SEQ ID No.2, and the downstream primer is shown in SEQ ID No.3. The specific company that produces the primers is Hangzhou Qingke Biotechnology Co., Ltd.
Embodiment 2
[0041] Establishment of Molecular Identification Methods
[0042] 1) DNA extraction of the sample to be tested
[0043] Pick a single colony of the strain to be tested in 5mL NA liquid medium, 180r min -1 , shake culture at 30℃ for 12h, until the concentration is 10 8 CFU mL -1 , centrifuged to collect the bacterial cells, using the TIANGEN Bacterial Genomic DNA Extraction Kit to extract the bacterial genomic DNA, and UV spectrophotometer Nanodrop 2000 to detect the quality and concentration of the extracted DNA. Standby at -20°C.
[0044] 2) Establishment of PCR amplification system and method
[0045] The PCR amplification reaction system is a 25 μL system, specifically: 1 μL DNA template, 1 μL upstream primer: Dda-F, 1 μL downstream primer: Dda-R, 12.5 μL Taq PCR Mix, 9.5 μL ddH 2 O.
[0046] The PCR amplification program was: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 10 s, and 35 cycles; exten...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com