Micro-RNA marker and application thereof
A marker and tiny technology, applied in the field of biomedical testing, can solve problems such as poor diagnosis and prognosis of CTEPH, achieve good tissue specificity, improve sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Primer Design
[0026] According to the sequence of hsa-miR-20a-5p, primers and probe sequences were designed. The specific sequence is as follows:
[0027] The reverse transcription primer sequence SEQ ID No.2 of hsa-miR-20a-5p:
[0028] GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT;
[0029] The forward primer sequence SEQ ID No.3 of hsa-miR-20a-5p:
[0030] GCGCGCTAAAGTGCTTATAGTGC;
[0031] The reverse primer sequence SEQ ID No.4 of hsa-miR-20a-5p:
[0032] GTGCAGGGTCCGAGGT;
[0033] The forward primer sequence of U6 is SEQ ID No.5: CTCGCTTCGGCAGCACA;
[0034] The reverse primer sequence of U6 is SEQ ID No.6: AACGCTTCACGAATTTGCGT;
[0035] The above primers were synthesized by GenScript Biotechnology Co., Ltd.
Embodiment 2
[0036] Example 2 total RNA extraction
[0037] (1) Take the blood sample (which has been added with Biotech lysate) and let it stand on ice for 5 minutes.
[0038] (2) Add 200 μl of chloroform, vortex and mix well, then place at room temperature for 2-3 minutes. Centrifuge at 12,000 g for 15 min at 4°C.
[0039] (3) Absorb the upper aqueous phase into another centrifuge tube. NOTE: Never pipette the middle interface.
[0040] (4) Add an equal amount of isopropanol, mix well, and precipitate at -20°C.
[0041] (5) Centrifuge at 12,000g at 4°C for 10 minutes, discard the supernatant, and sink the RNA to the bottom of the tube.
[0042] (6) Add 1ml of 75% ethanol to suspend the precipitate.
[0043] (7) Centrifuge at 12,000 g for 5 min at 4°C, and discard the supernatant as much as possible.
[0044] (8) Air dry at room temperature or vacuum dry for 5-10min. Note: RNA samples should not be too dry, otherwise it will be difficult to dissolve.
[0045] (9) with 20μl DEPC H ...
Embodiment 3
[0047] Example 3 Prediction of miRNAs associated with CTEPH
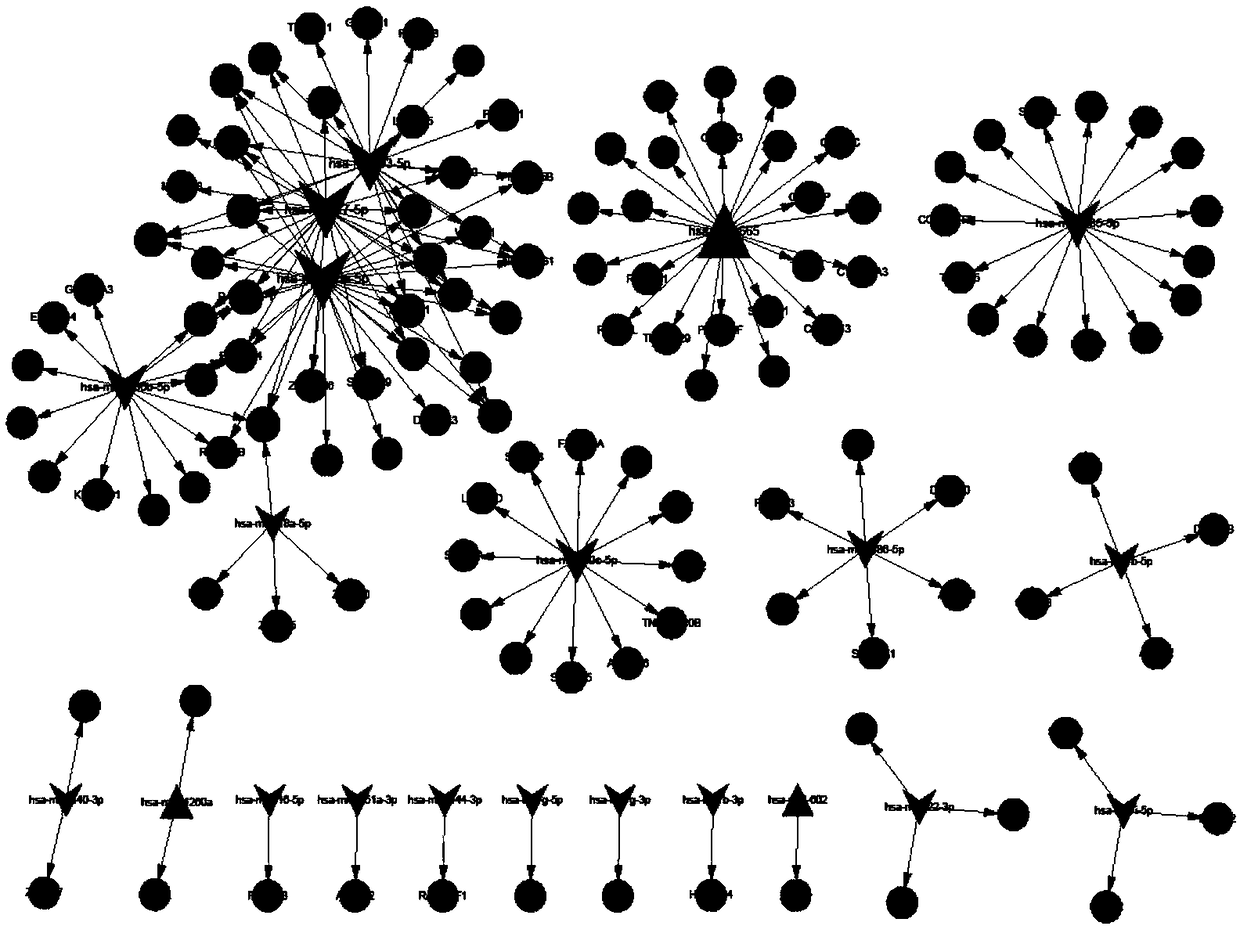
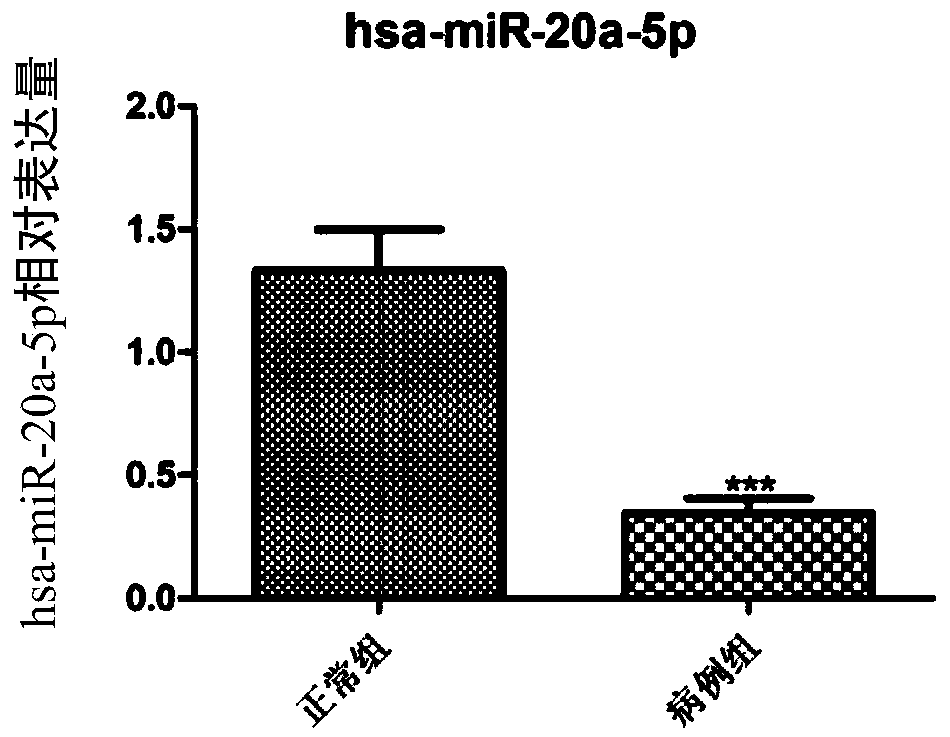
[0048] Using bioinformatics analysis technology, 169 miRNA-target relationship pairs were predicted by miRWalk2.0 software, including 21 miRNAs, 3 up-regulated miRNAs, and 18 down-regulated miRNAs; the above predicted miRNA-target relationship pairs were used to construct a regulatory network Figure, miRNA-target regulatory network see figure 1 , a total of 147 nodes, see the specific results figure 1 . Among them, hsa-miR-20a-5p is a down-regulated miRNA associated with CTEPH disease.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com