A method for mitochondrial sequence assembly and copy number determination based on high-throughput sequencing
A technology of sequence splicing and mitochondria, applied in the field of genomics, can solve the problems of complicated operation, high sample quality requirements, and troublesome design, and achieve the effect of simple experiment, low sample quality requirements, and low unit cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
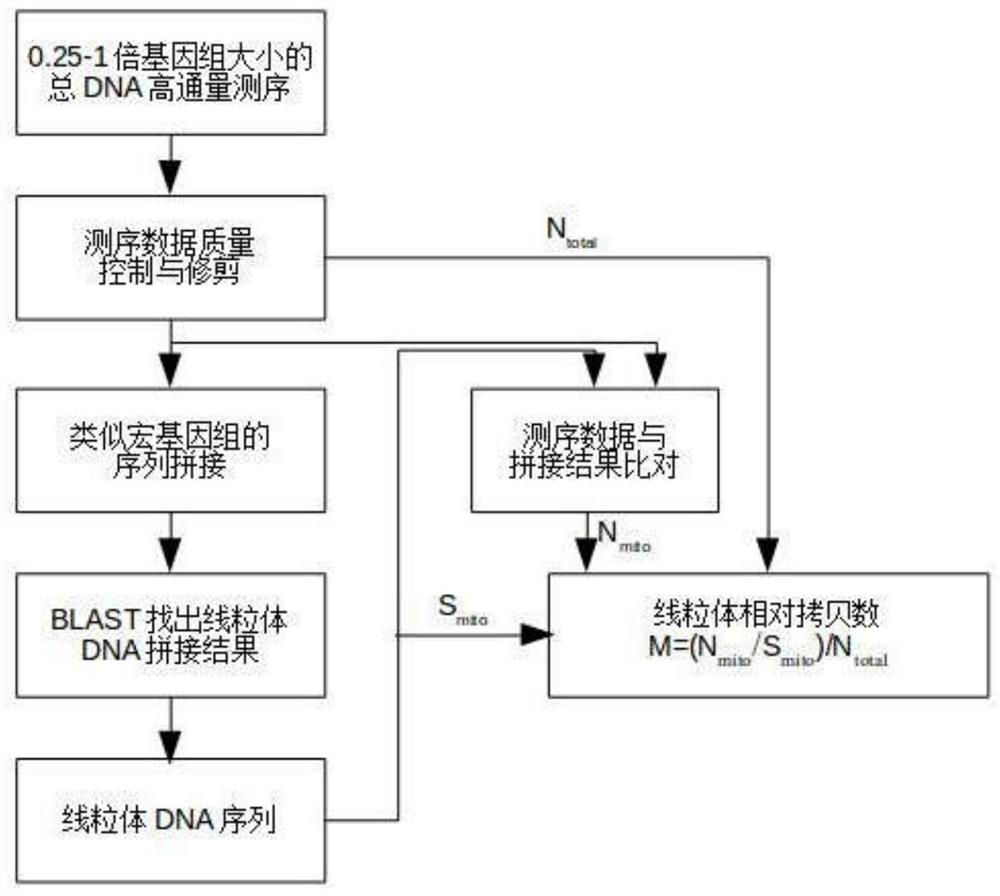
[0036] Mitochondrial DNA sequencing and stitching three cases Mitten, comprising the steps of:
[0037] 1. Total DNA 3 Chinese mitten only muscle tissue, sonicated to break 500-700bp, using DNA library construction kits construct a high-throughput sequencing library.
[0038] 2. Construction of high-throughput sequencing and library use NextSeq500 high-throughput sequencing sequencing, sequencing each sample volume of 2G. And use the software bcl2fastq sequencing results into fastq format.
[0039] 3. Trimmomatic software sequencing data for quality control, particularly for the key parameters: LEADING: 5TRAILING: 5SLIDINGWINDOW: 4: 15MINLEN: 30, to give high quality sequencing data.
[0040] 4. SPAdes software sequencing data of the above-described stitching quality, particularly for the key parameters: - meta-k 55 (see Table 1).
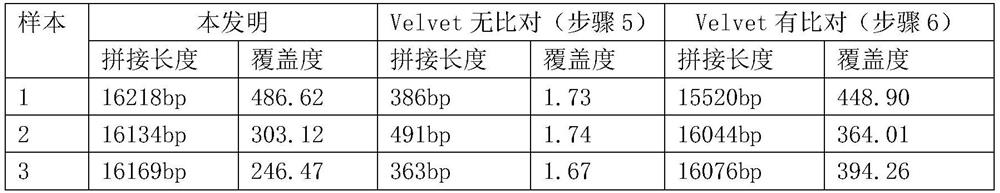
[0041] 5. The control group of the above-described velvet software sequencing data quality stitching, specifically the key parameters: -cov_cutoff 3...
Embodiment 2
[0048] The effects of different sequencing amounts and mitochondrial DNA sequence occupy the comparison of splicing effects, the steps are as follows:
[0049] 1. Simulate data of different sequence of sequence and mitochondrial DNA sequence ratio. Using high quality sequencing data in Example 1, the data of 1 / 3, 1 / 6 and 1 / 12, and using data without mitochondria DNA with the aforementioned randomly selected data, to total data 2G, 1G and 0.5g.
[0050] 2. Use the Spades software to splicing the analog data, and the splicing process will be operated in step 4 in the above embodiment 1.
[0051] 3. Find the mitochondrial DNA sequence of splicing in the splicing result, the method is the same as in the first embodiment.
[0052] The splicing results of the simulation results are as follows:
[0053] sample Data Size (BP) Sample 1 sequence occupation Splicing length (BP) KMER coverage E1 2G 1 / 3 16096 127.98 E2 1G 1 / 6 16096 64.34 E3 0.5G 1 / 12 16091 ...
Embodiment 3
[0056] The calculation of mitochondrial copy numbers in sequencing data, the steps are as follows:
[0057] 1. Sequencing and splicing data are data and splicing results used in Example 1. Use the ReseqTools software to view the total base base of high quality data, remember to n total .
[0058] 2 calculate the total length of mitochondrial DNA after stitching, remember to mito .
[0059] 3 Use Bowtie2 software to bring high quality data to the mitochondrial splicing result sequence and generate SAM data files. The above SAM file is calculated using the FlagStat function of SamTools to obtain the total base of the total base of READs that can be compared to the mitochondrial DNA sequence. mito .
[0060] 4 Calculate the normalization copy number m = (n mito / S mito ) / N total . The results are as follows:
[0061] sample N total (bp)
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com