Application of soybean E3 ubiquitin ligase family gene GmRNF1a
A soybean, coding gene technology, applied in the direction of ligase, application, genetic engineering, etc., can solve problems such as different functions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Cloning and identification of soybean GmRNF1a and its coding gene
[0032]Primers were designed according to the sequence information of GmRNF1a predicted by the phytozome website, and the flower cDNA of Willmas82 in full flowering stage was used as a template for PCR amplification.
[0033] Upstream primer GmRNF1a-F: ggatcttccagagatGCTTCTTCACTTCTTCCATTCTCC; (SEQ ID NO.3)
[0034] Downstream primer GmRNF1a-R: ctgccgttcgacgatCCTTATTGTAAACGTCGTTATCAGC. (SEQ ID NO.4)
[0035] The GmRNF1a gene was amplified from the total RNA of soybean floral organs by RT-PCR. Take the soybean flower tissue, grind it with a mortar, add it into a 1.5mL EP tube filled with lysate, shake it fully, and then transfer it into a glass homogenizer. After homogenization, transfer to 1.5mLEP tube, and use plant total RNA extraction kit (TIANGEN DP404) for total RNA extraction. The quality of total RNA was identified by formaldehyde denaturing gel electrophoresis, and then the RNA conte...
Embodiment 2
[0036] Example 2 Expression characteristics of GmRNF1a in different organs of soybean
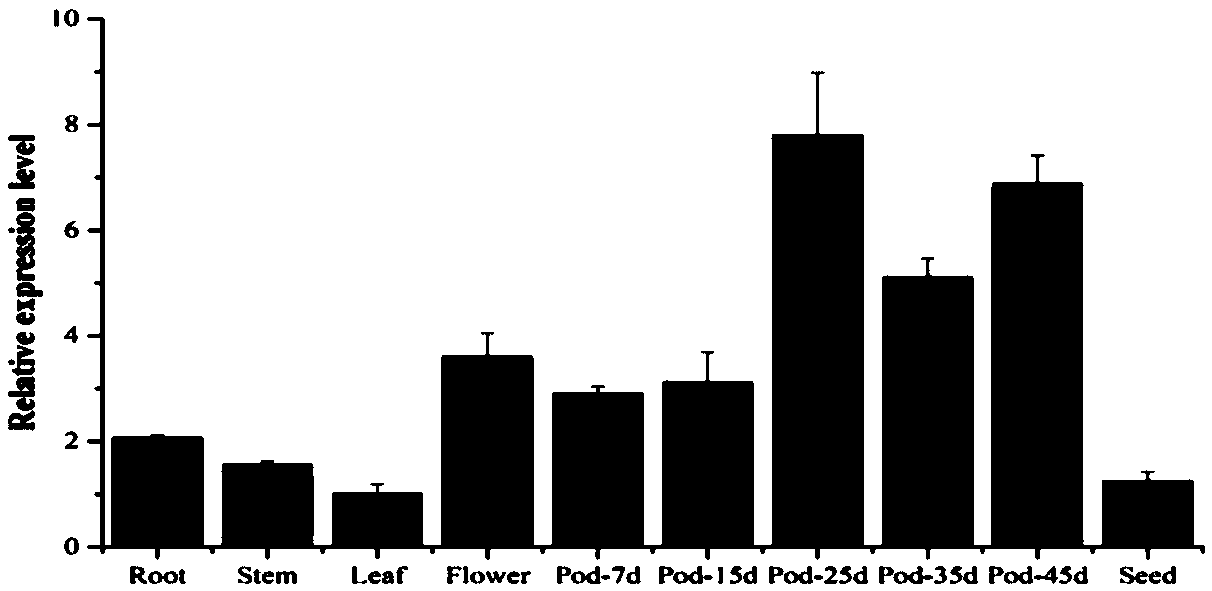
[0037] Extract RNA from roots, stems, leaves, flowers, 7d pods, 15d pods, 25d pods, 35d pods, 45d pods and seeds of Willmas82, reverse to cDNA for RT-PCR analysis.
[0038] The extraction of total RNA was the same as in Example 1. The soybean constitutively expressed gene Tubulin was used as an internal reference gene, and the amplification primers were Tubulin forward primer sequence: GGAGTTCACAGAGGCAGAG (SEQ ID NO.5), and Tubulin reverse primer sequence: CACTTACGCATCACATAGCA (SEQ ID NO.6). Using cDNA from different soybean tissues or organs as templates, real-time fluorescent quantitative PCR analysis was carried out. The amplification primers of GmRNF1a are: GmRNF1a-qPCR-F: CCGTGGATAGAACTCAACTCG (SEQ ID NO.7), GmRNF1a-qPCR-R: GTCCTGAGCCCGTAGAAATC (SEQ ID NO.8). result( figure 2 ) analysis showed that the expression of GmRNF1a in flowers and pods was relatively high, especially in the...
Embodiment 3
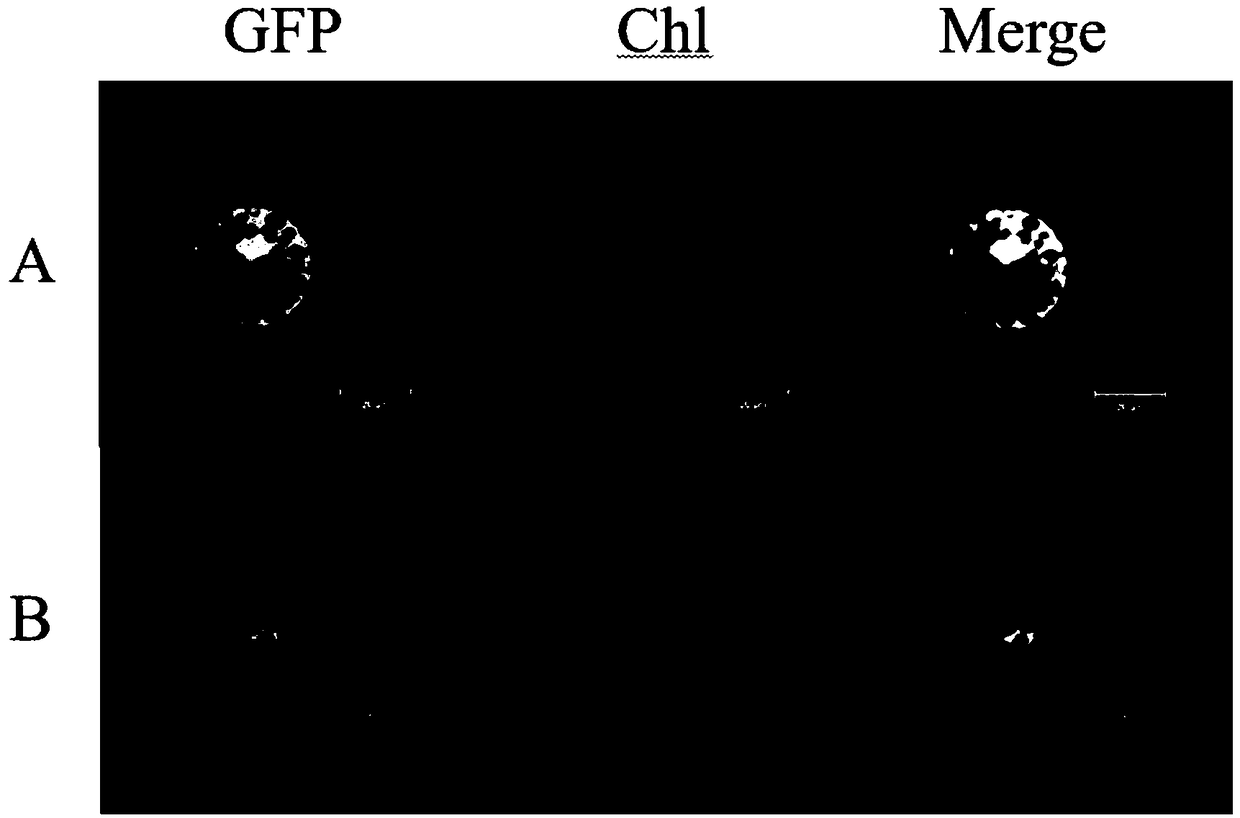
[0039] Example 3 Subcellular localization of GmRNF1a
[0040] Subcellular localization adopts the method of transient expression of Arabidopsis protoplasts, the vector used is pAN580, and the primers are GmRNF1a-AN-F: aagtccggagctagctctagATGGCGGCGACGGCGACG (SEQ ID NO.9), GmRNF1a-AN-R: gcccttgctcaccatggatccGCAAGCCGAATCGCCGCCA (SEQ ID NO.10 ). PCR amplification, after the target band is correct, it is recovered by tapping the rubber, and the recovered product of the rubber is connected to the carrier by homologous recombination to construct the subcellular localization vector pAN580-GmRNF1a (the gene is at the N-terminal of GFP), and Arabidopsis protoplasts are transiently After expression, it was cultured in the dark for 16 hours, and after laser irradiation by a laser confocal microscope (Zeiss, LSM780), a green fluorescent signal could be generated, the protein was located, observed and photographed. The result is as Figure 3-5 As shown, the transformed empty plasmid is di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com