Gene detection kit including nucleic acid molecules of universal fluorescent subtracted probe and application thereof
A detection kit and gene detection technology, applied in the field of gene detection, can solve the problems of complicated operation, difficult to popularize in a large area, and difficult to standardize hybridization conditions.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Embodiment 1 Gene detection method of the present invention
[0074] 1. The composition of the detection kit
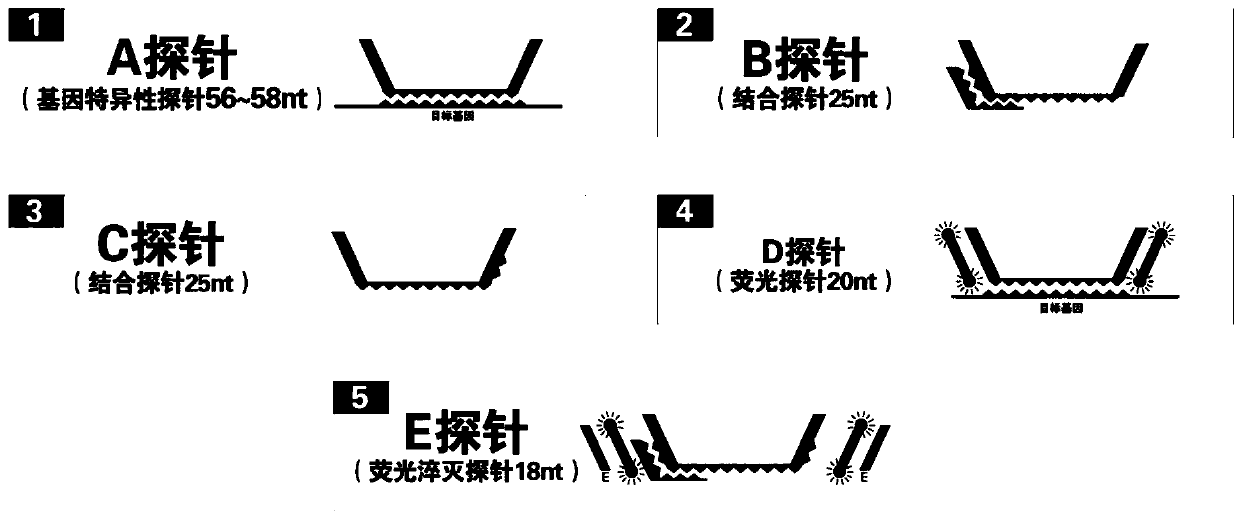
[0075] Schematic as figure 1 Shown:
[0076] A sequence:
[0077] 5′–CTGAAGCGCTTCGCGAGCCM 1 m 2 N 1 N 2 N 3 N 4 N 5 …N 6 N 7 N 8 N 9 N 10 m 3 m 4 CTGAAGCGCTTCGCGAGCC-3'; wherein, N 1 N 2 N 3 N 4 N 5 …N 6 N 7 N 8 N 9 N 10 is the complementary sequence of the gene to be tested or its fragment, M 1 m 2 and M 3 m 4 is the linker sequence;
[0078] B sequence: 5′-n 5 no 4 no 3 no 2 no 1 m 2 m 1 TAGGCTCGCGAAGC-3'; n1 no 2 no 3 no 4 no 5 with N 1 N 2 N 3 N 4 N 5 The sequence complement of m 1 m 2 with M 1 m 2 sequence complementary;
[0079] C sequence: 5′-GCGAAGCGCTTCAGm 4 m 3 no 10 no 9 no 8 no 7 no 6 -3'; n 6 no 7 no 8 no 9 no 10 with N 6 N 7 N 8 N 9 N 10 The sequence complement of m 3 m 4 with M 3 m 4 sequence complementary;
[0080] D sequence: 5′.-FAM-GGCTCGCGAAGCGCTTCAG–FAM-3′
[0081] E ...
Embodiment 2
[0089] Embodiment 2 Gene detection method of the present invention
[0090] 1. The composition of the detection kit
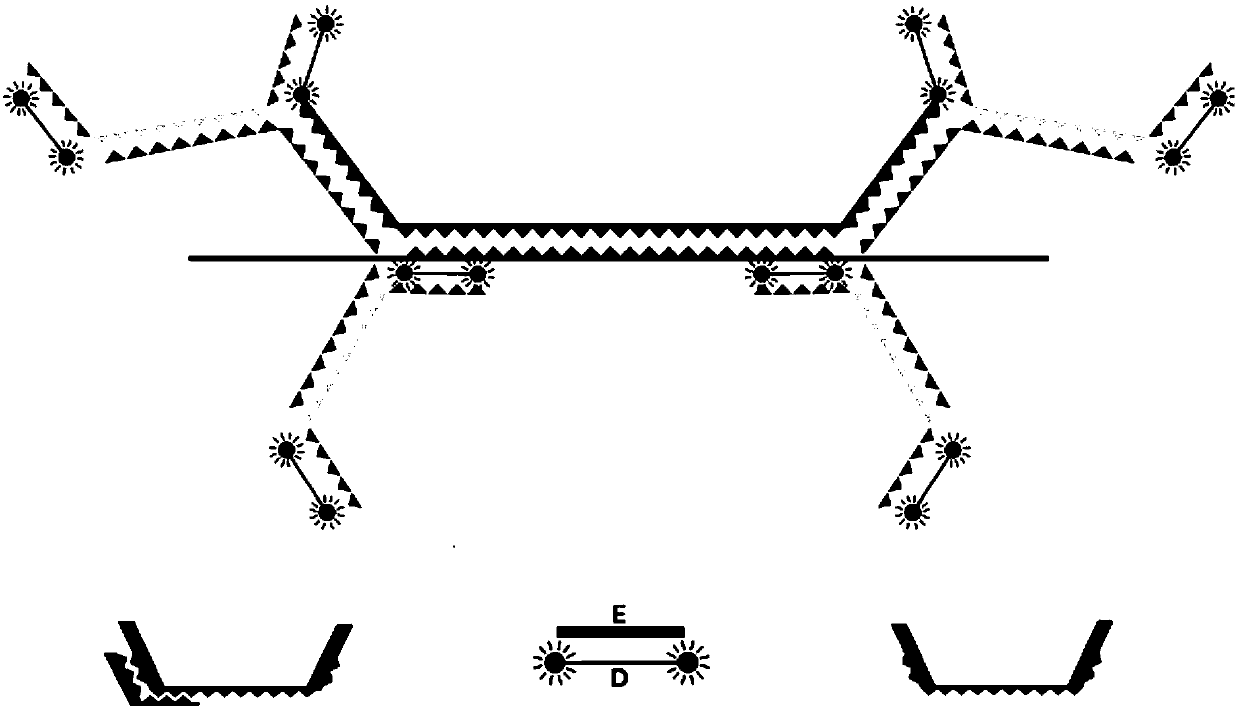
[0091] Schematic as figure 2 Shown:
[0092] ⑴A sequence:
[0093] 5'–CTGAAGCGCTTCGCGAGCCM 1 m 2 N 1 N 2 N 3 N 4 N 5 …N 6 N 7 N 8 N 9 N 10 m 3 m 4 CTGAAGCGCTTCGCGAGCC-3'; wherein, N 1 N 2 N 3 N 4 N 5 …N 6 N 7 N 8 N 9 N 10 is the complementary sequence of the gene to be tested or its fragment, M 1 m 2 and M 3 m 4 is the linker sequence;
[0094] Among them, M 1 m 2 for TA or AC, and / or, M 3 m 4 It is AT, CA or TC.
[0095] ⑵B sequence: 5'-n 5 no 4 no 3 no 2 no 1 m 2 m 1 TAGGCTCGCGAAGC-3'; n 1 no 2 no 3 no 4 no 5 with N 1 N 2 N 3 N 4 N 5 The sequence complement of m 1 m 2 with M 1 m 2 sequence complementary;
[0096] CDC sequence: 5'-GCGAAGCGCTTCAGm 4 m 3 no 10 no 9 no 8 no 7 no 6 -3'; n 6 no 7 no 8 no 9 no 10 with N 6 N 7 N 8 N 9 N 10 The sequence complement of m 3 m 4 with M 3 m 4 se...
experiment example 1
[0123] Experimental example 1 adopts the method of the present invention to detect brain cell edema gene AQP4 gene
[0124] 1. Detection method
[0125] The sample to be tested is the brain tissue nerve cells of rats with cerebral edema.
[0126] The inventive method detects according to the method of embodiment 1:
[0127] (1) First, design and synthesize a set of fluorescence subtraction probes according to the mRNA sequence of the AQP4 gene, and its sequences and labels are respectively:
[0128] A sequence:
[0129] 5′-.CTGAAGCGCTTCGCGAGCCTAGCTACATGGAGGTGGAGGACAACCGATCTGAAGCGCTTCGCGAGCC-3′
[0130] Sequence B: 5′.-CCATGTAGCTAGGCTCGCGAAGC-3′
[0131] C sequence: 5'.-GCGAAGCGCTTCAGATCGGTTGTC-3'
[0132] D sequence: 5′.-FAM-GGCTCGCGAAGCGCTTCAG–FAM-3′
[0133] E sequence: 5'-.TAMRA-CTGAAGCGCTTCGCGAGC-TAMRA-3';
[0134] (2) Dissolve the above-mentioned probes in TE buffer respectively, so that the concentration of sequence A is 1umol / L, the concentration of sequence B is...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com